| While the main mechanism of turning genes
on and off occurs at the level of transcription, the amount
of active gene produced can also be regulated by events
occurring after transcription. In fact, we have already
covered (and you have been tested on) some of these
processes under RNA processing on a previous outline. (I
included the topics below in small type
just as a reminder). By way of RNA processing, the
ultimate expression of the gene is altered. However,
there are also other post-transcriptional processes we
have not yet covered (in normal size type below). |
|
|
- Translational Regulation
of Gene Expression: The expression of a gene
can also be regulated at the level of translation.
- mRNA
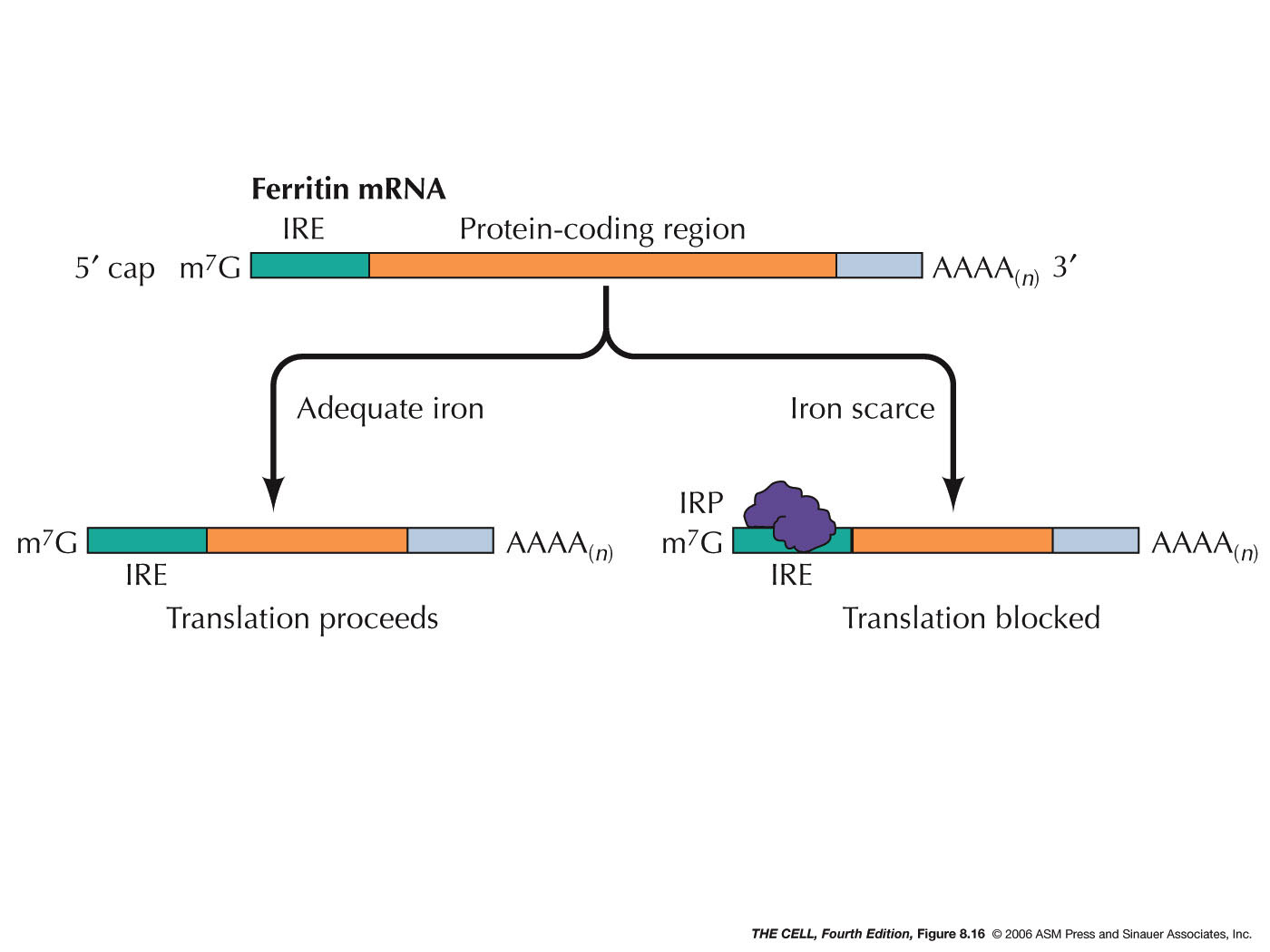
Inactivation by Protein Binding: A protein
can bind to a specific mRNA and thereby block its
translation, as occurs in ferritin synthesis.
This protein is an intracellular iron-storing molecule
and therefore the higher the iron concentration in the
cell, the more ferritin is needed. (Ferritin is also
found in lower concentration in plasma.) Regulation is
due to the binding of an iron regulatory protein
(IRP)(IRP1 and IRP2) to the iron response element
(IRE) of the ferritin mRNA. This site is in the 5' UTR
(untranslated region) of the mRNA. This binding
prevents the small ribosomal subunit from scanning for
the AUG initiator codon, thereby turning off
translation. In the presence of iron, IRP1 become
inactive (does not bind to IRE) and IRP2 is degraded
and translation can proceed. Other proteins bind to
the 3' UTR and act as translation
repressors.
|
|
|
|
 |
- Polyadenylation
Changes: A protein binds to some oocyte mRNAs
and shortens their 3' poly-A tails from about 200 nt
to about 30-50 nt. This causes them to be inactive
before fertilization. They become translationally
active at the right stage of development due to
re-polyadenylation.
|
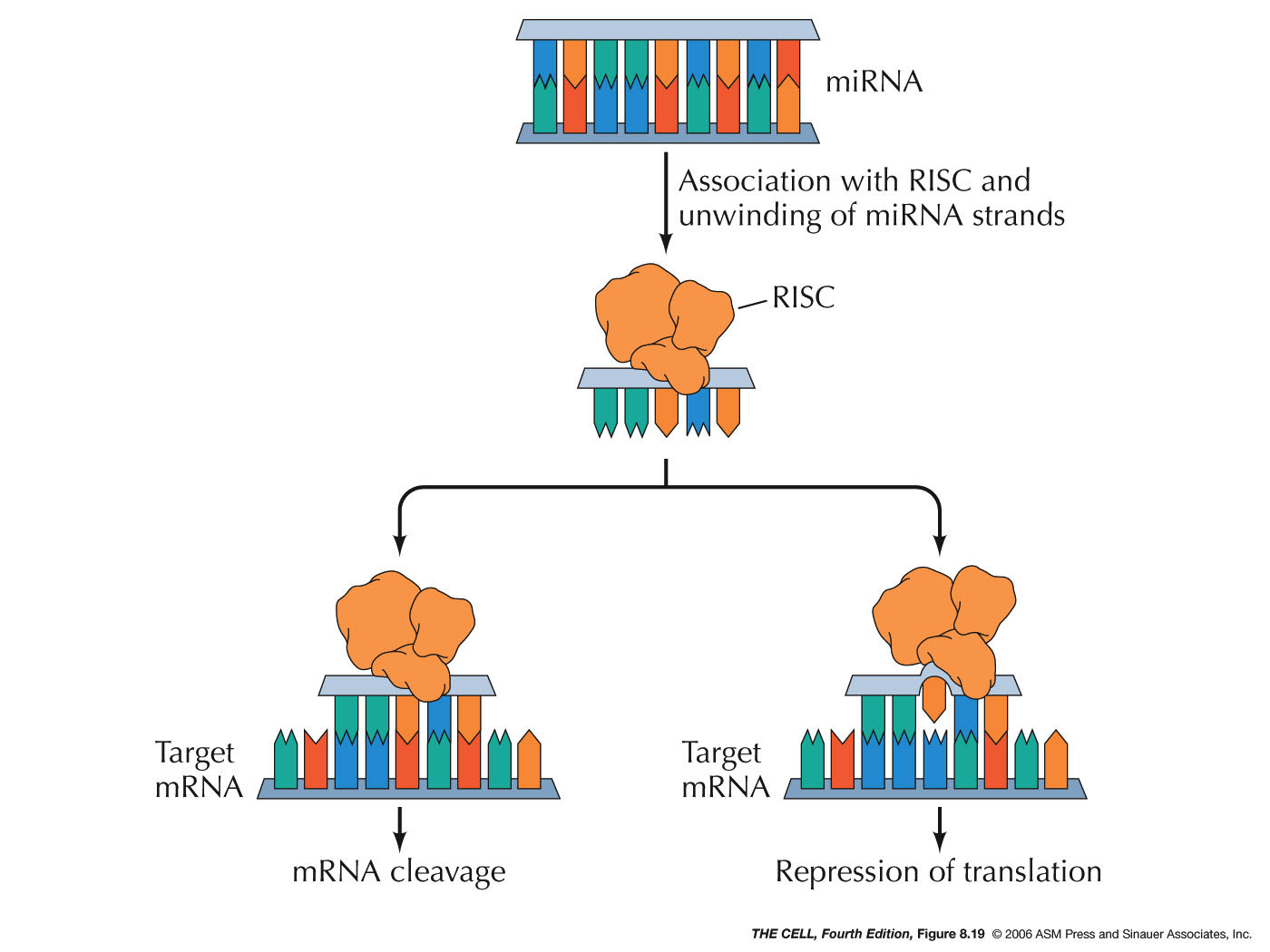
- miRNAs
and siRNAs: MicroRNAs (miRNAs) and
small interfering RNAs (siRNAs) can down-regulate the
translation process. These RNAs may come from
processing of intron RNA or from mRNA UTRs. Others are
transcribed as a larger RNA that can be processed to
produce more than one miRNA (or siRNA). An RNase
called dicer converts larger molecules into a double
stranded short RNA molecule. The final products
(miRNAs and siRNAs) are about 20-26 nucleotides long
and seem to be involved in the regulation of about one
third of all human genes (over 1000 miRNAs have been
discovered in humans and over 1.5 million in Arabidopsis!).
One strand of the RNA then binds to a protein
called Argonaut.
This complex is called the RNA-induced silencing
complex (RISC). This RISC complex then can bind to the
3' UTR of an mRNA. If this complementary base pairing
is precise (siRNAs), it stimulates the cleavage of the
mRNA. If it is not precise (miRNAs), it has the effect
of inhibiting translation. (Small RNAs can also
inhibit transcription.)(miRNAs
and
cancer and here)(miRNAs
and immunity)(siRNA
vs. miRNA)
- Hunting for
miRNAs: Genes for these small,
non-transcribed RNA are hard to find. One reason
is that cells or organisms with a lack of an
miRNA may not have a clear phenotype. Evidence
from Caenorhabditis elegans indicate
that there is miRNA redundancy and knocking out
one miRNA may only produce a phenotype when the
worm is under physical stress (Victor Ambros
et al., GSA 2012)
|
 |
- Post-translational
Regulation of Gene Expression: Even after the
protein is produced, it effectiveness (expression of the
gene) can be modulated. This process occurs by way of
several process we will take up later as we study the
cytoplasm in more detail (protein folding and chaperons,
disulfide bridge formation, protein cleavage, protein
glycosylation, allosteric regulation, protein
phosphorylation, protein degradation and others).
- Inteins:
These are amino acids sequences that have the ability
to splice themselves out of a polypeptide. They have
also been called protein introns.
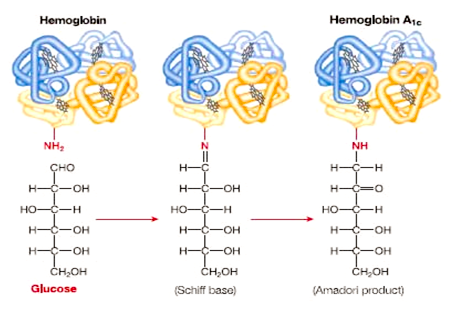
-
Hemoglobin A1c:
While this example of a protein that is modified
after translation has nothing to do with the
expression of the gene (that is, it is not related
to whether or not the protein is active), it is a
case of protein modification that is useful in
disease diagnosis. Patients that develop diabetes 2
have traditionally been diagnosed by elevated blood
glucose levels, but since these levels can vary with
periods of fasting and carbohydrate consumption, an
easier, more reliable test was needed. As it turns
out, hemoglobin that has been exposed to high levels
of glucose becomes chemically modified into Hb A1c.
Since the half life of a red blood cell is about 3
months, the degree of modification (glycation) of
hemoglobin can be used a an indicator of the average
glucose level over the last three months. A level of
Hb A1c ≥ 48 mmol/mol is an indication of diabetes
(American Diabetes Association). The reaction in
which a sugar is added to the N-terminus of the
β-chain of hemoglobin is shown here
(http://usmle.biochemistryformedics.com/hyperglycemia-induced-complications-in-diabetes-mellitus/).
|
|
 Home
Home



