|
Molecules
The molecules of the cell are classified as
inorganic compounds (relatively small with little or no
carbon) or organic compounds (larger molecules rich in
carbon).
Inorganic:
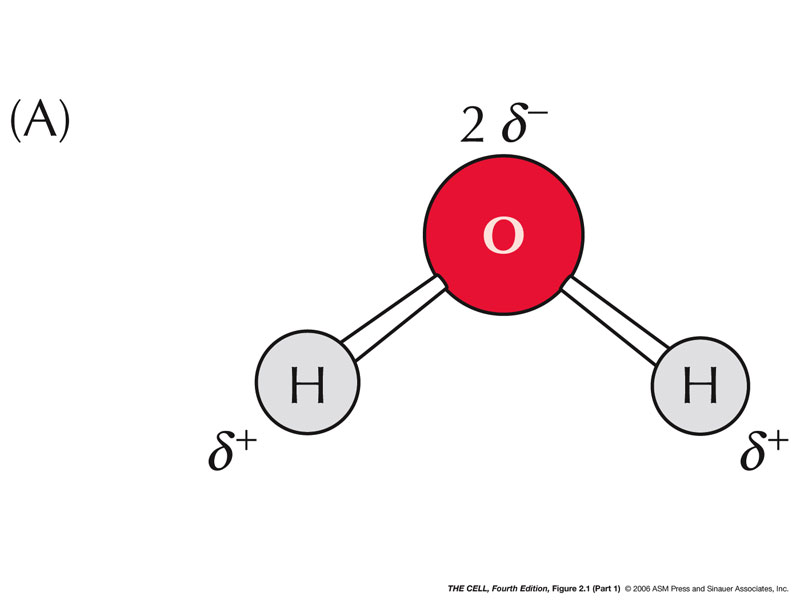
- Water:
Water is the most abundant molecule in cells (~70% in
the average cell). It is a polar molecule and the
intermolecular hydrogen bonds are responsible for many
of its properties. It is an excellent solvent.
- Other:
Numerous other inorganic, such as various ions,
dissolved gases, and others, are important in many
cellular mechanisms.
|
 |
|
Organic:
- Carbohydrates:
These molecules have abundant chemical potential energy.
They are called carbohydrates because the atoms C:H:O
are approximately in a 1:2:1 ratio (carbo = C; hydrate =
water).
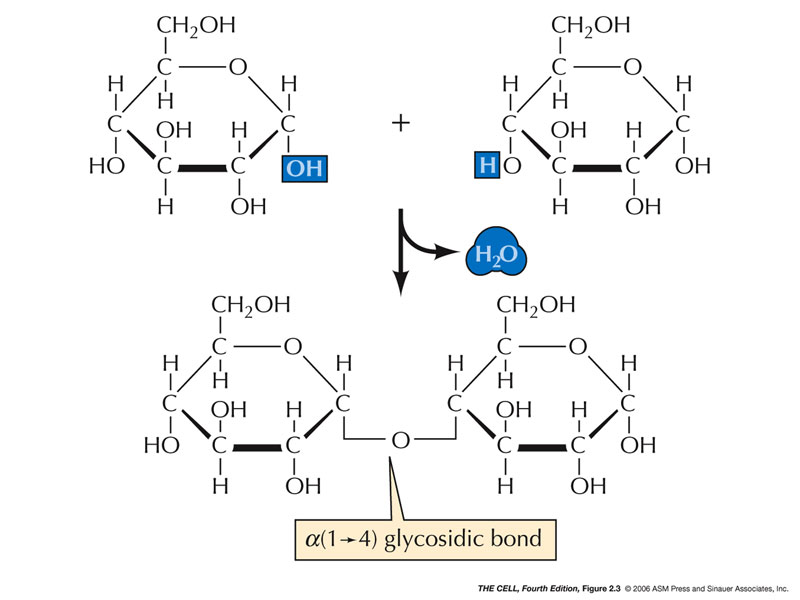
- Sugars:
Sugars are smaller carbohydrates and may be linear or
ring shaped.
- Monosaccharides (Simple Sugars):
These are 3-7 carbons carbohydrates. Often they have
5 or 6 carbons.
|
 |
 |
- Polysaccharides/Oligosaccharides:
These carbohydrates are composed of 100s to 1000s of
monosaccharides.
- Starch (contrary
to what this movie says, starch is not in animals): This class of
carbohydrates is found in plants; amylopectin is an
example and has some alpha (1-6) bonds making it a
branched molecule.
- Glycogen:
This is a large carbohydrate found in animals.
(Sometimes called "animal starch.") Also branched.
- Cellulose:
This carbohydrate is present in plant cell walls and
has beta (1-4) bonds (we cannot digest them). (Dietary
fiber
and colon cancer.)(Dietary
fiber
and diabetes)
|
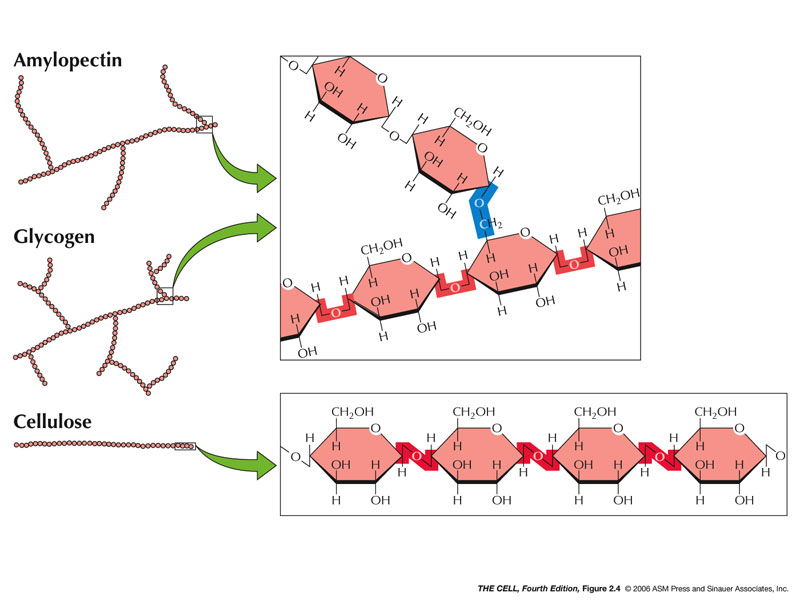 |
- Glycoproteins/Glycolipids:
carbohydrate-protein, carbohydrate-lipids)
|
 |
- Lipids:
These are also energy molecules, some involved in
cell signalling.
- Fatty Acids:
These are hydrocarbon(Fatty acids may be as short as 4
Cs, or as long as 28 Cs and most naturally-occurring
fatty acids have an even number of Cs, according to
that infallible site, Wikipedia. Many have 16 or 18 Cs
or somewhere in the low 20s.)
- Saturated Fatty Acids: Saturated
fatty acids have no double bonds between the carbon
atoms (saturated with hydrogens).
- Unsaturated Fatty
Acids: These have at least one
double bond.
- Monounsaturated Fatty Acids: These
have exactly one double bond.
- Polyunsaturated Fatty Acids: These
have more than one double bond and are good for
cardiovascular health.
|
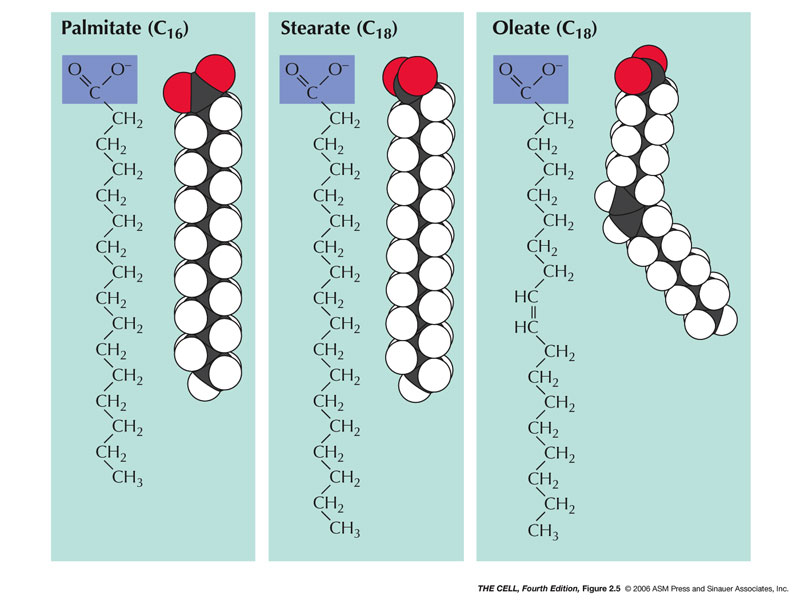 |
- Trans
Fats versus Cis Fats: Recent
discoveries have shown that the configuration
around the double bond is extremely important:
trans = bad and cis = good for cardiovascular
health. (Trans
fats
and heart disease)(Ranking
of fatty acids according to heart health)
|
 |
- Triacylglycerols
(triglycerides, fats): These lipids are
composed of a glycerol plus 3 fatty acids.
- Glycerol:
A 3 carbon molecule with 3 hydroxyls.
- Fatty Acids: A hydroxyl of glycerol
reacts (synthesis reaction) with the hydroxyl of the
carboxyl group of the fatty acid forming an ester
bond.
|
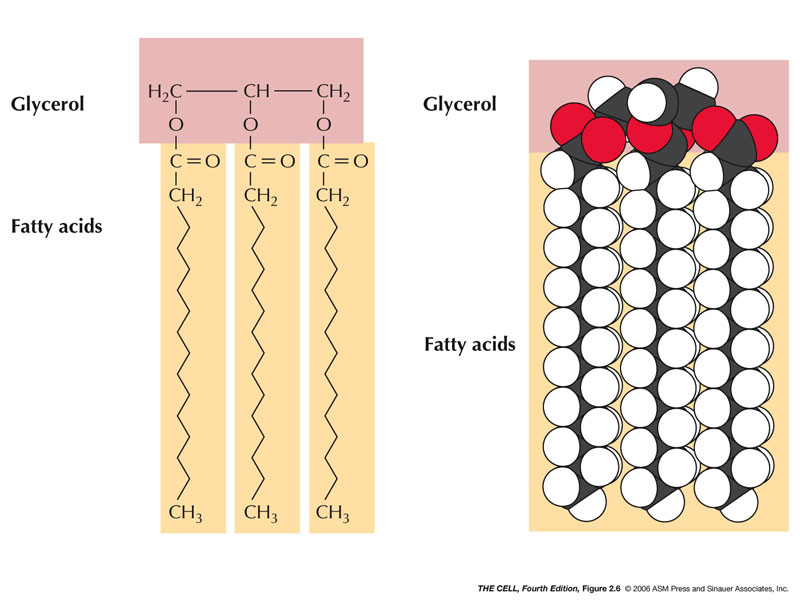 |
- Phospholipids:
These are common membrane lipids and are amphipathic.
- Phospholipid: This is a glycerol + 2
fatty acids + another phosphate-containing small
molecule.
- Sphingomyelin: This is similar to
phospholipids but has serine + 2 fatty acids.
|
 |
- Glycolipids:
These are also amphipathic made up of a carbohydrate +
another molecule + 2 fatty acids.
|
 |
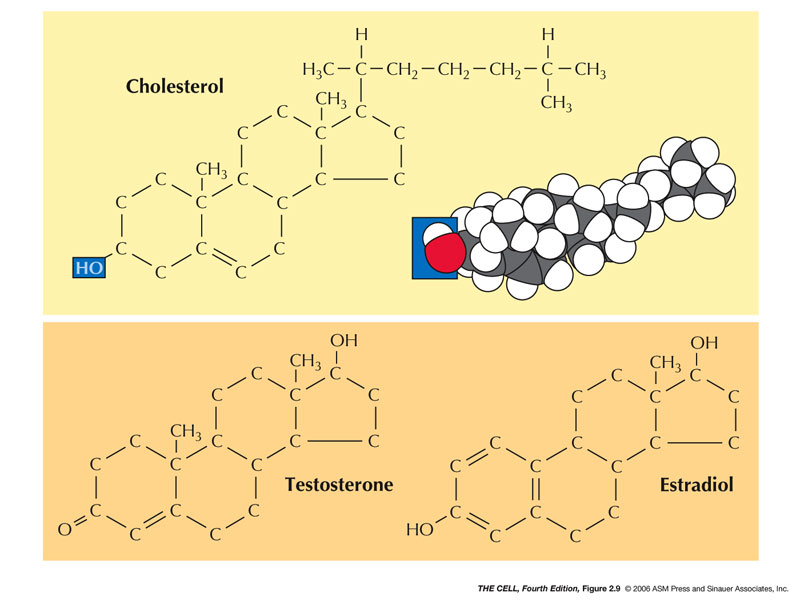
- Steroids:
These molecules have hydrocarbon rings (4) with a
hydroxyl and also are amphipathic.
- Cholesterol:
A common membrane steroid.
- Steroid Hormones:
Many (but not all) hormones are steroids, including
cortisol and testosterone.
|
 |
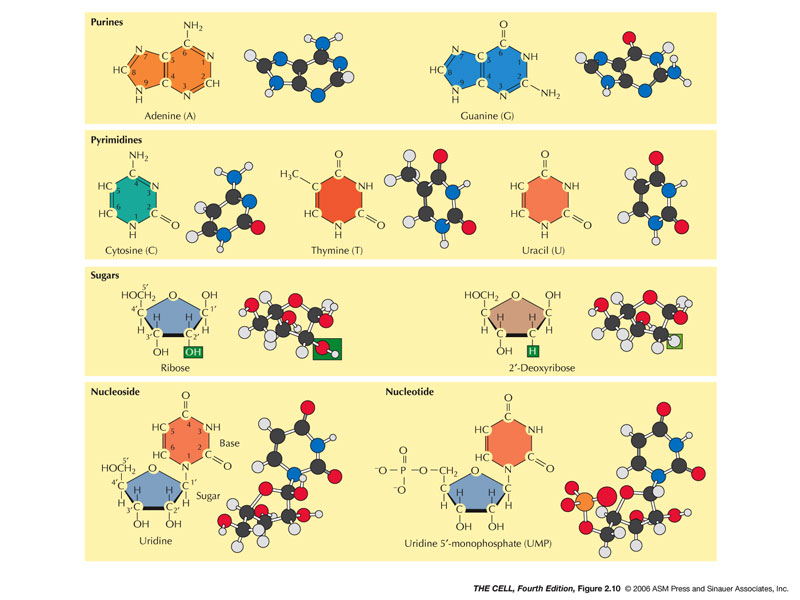
- Nucleic Acids
(and Related Molecules): These are
polynucleotides. There are two types: DNA and RNA.
A nucleotide is composed of a phosphate, a pentose
sugar, and a nitrogen base.
- Nucleotides: These
are
the building blocks of polynucleotides but they also
include other molecules important molecules like ATP
and cAMP.
- Pentose: The pentose of RNA is
ribose and the pentose of DNA is deoxyribose
(2'-deoxyribose).
- Nitrogen Base (Base): Nitrogen bases
are ring-molecules and come in two basic shapes:
purines and pyrimidines.
- Purines:
Purines are double-ring molecules.
- Adenine,
Guanine: These are
the two types of purine and are found in both
DNA and RNA.
- Pyrimidines:
These are single ring molecules.
- Cytosine,
Thymine, Uracil:
Cytosine is found in DNA and RNA; thymine is
found only in DNA; uracil is found only in
RNA.
|
 |
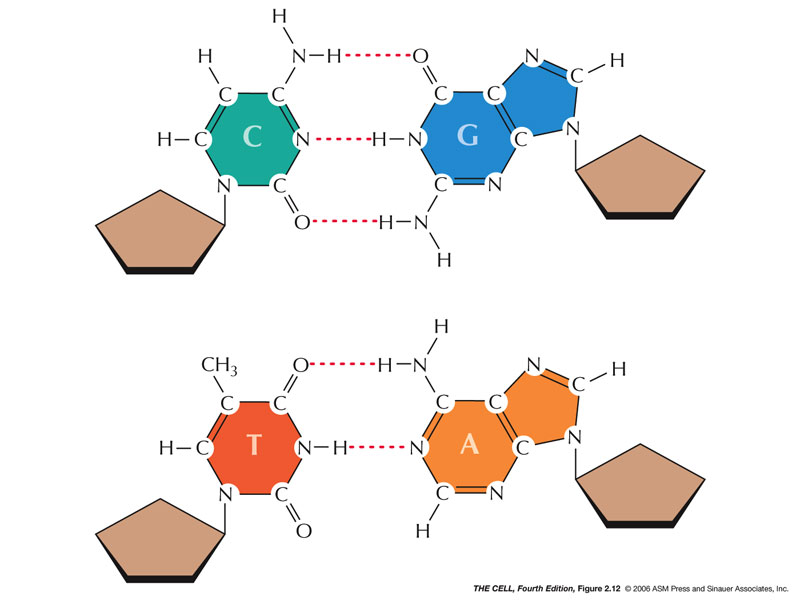
- Base Pairing:
Hydrogen bonds:
A-T (or A-U) and G-C pairs base pairs form between the
bases of polynucleotides.
- Nucleosides
versus
Nucleotides: A nucleoside is a sugar-base. A
nucleotide is a sugar-base-phosphate (may have more
than one phosphate).
|
 |
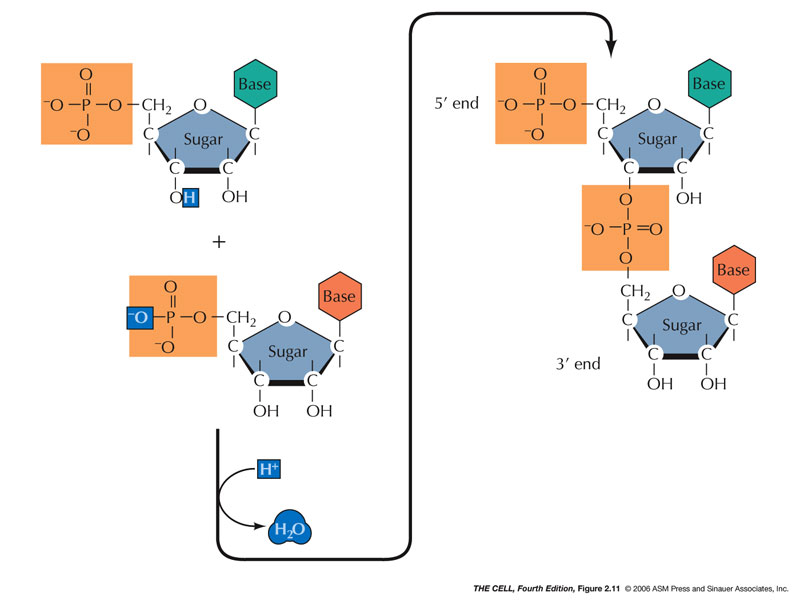
- Polynucleotides: DNA and RNA:
DNA and RNA have polarity; they have 5' and 3' ends.
|
 |
- Proteins:
Proteins are polypeptides and have numerous functions
including structure, defense, and enzymes.
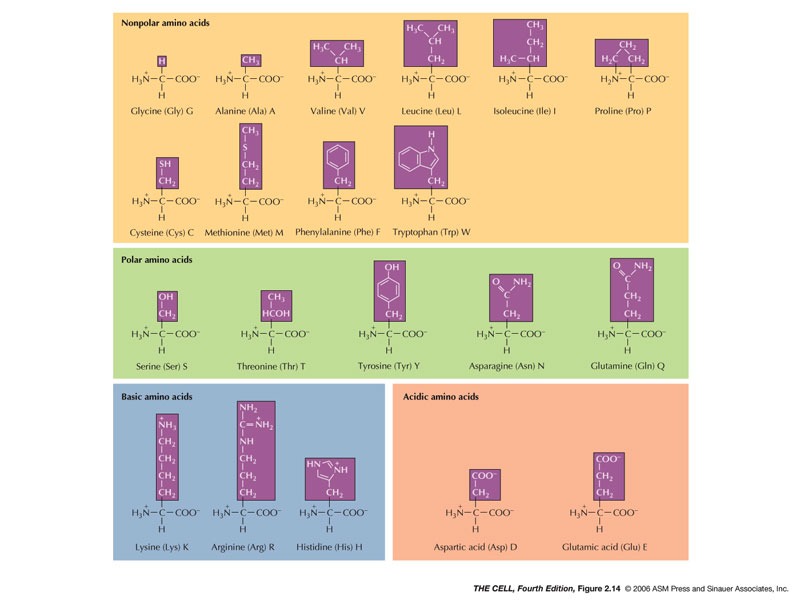
- Amino Acids:
There are 20 kinds of amino acids. Each has an amino
end and a carboxyl end with a unique radical. The
radical gives the amino acid it characteristics:
|
 |
|
|
 |
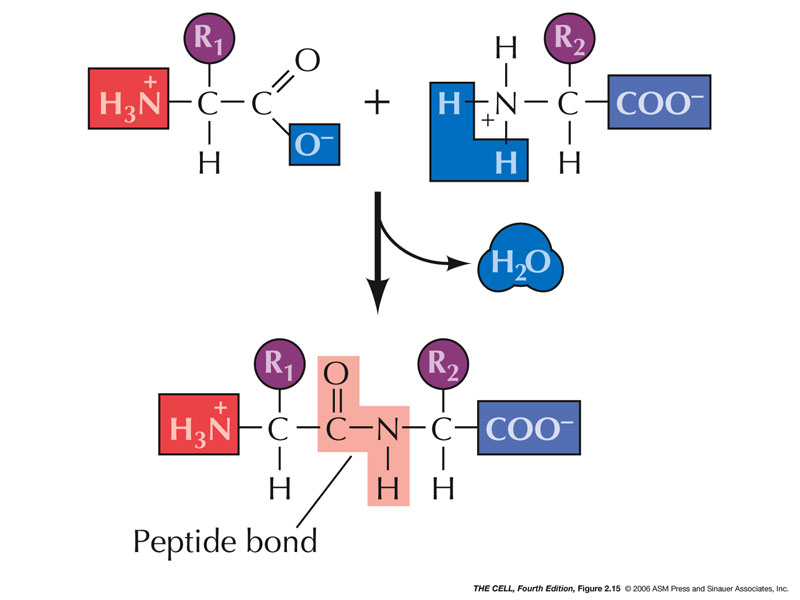
- Peptide Bond:
The carboxyl end of one amino acid forms a covalent
bond with the amino end of another amino acid
(synthesis reaction): the C-N peptide bond.
- Protein Structure:
Proteins have three or four "degrees of structure."
- Primary
Structure: This
is the amino acid sequence and is all important.
|
 |
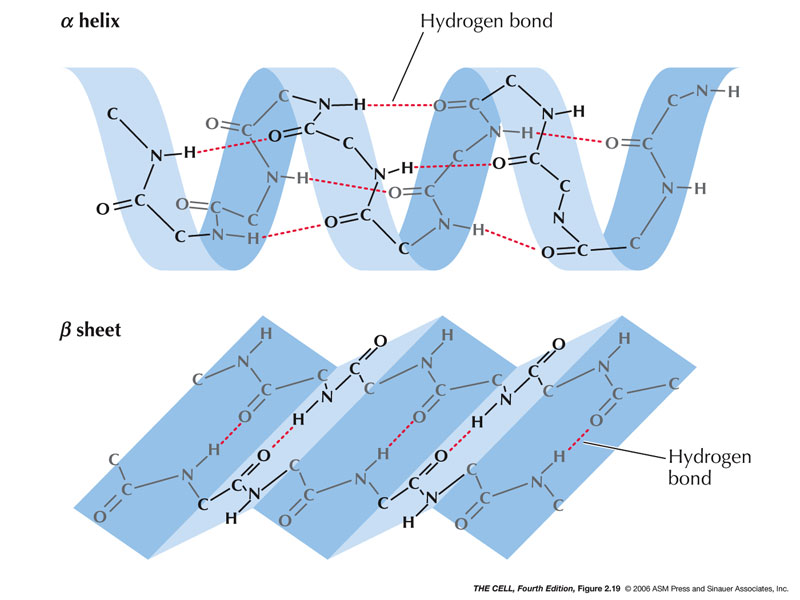
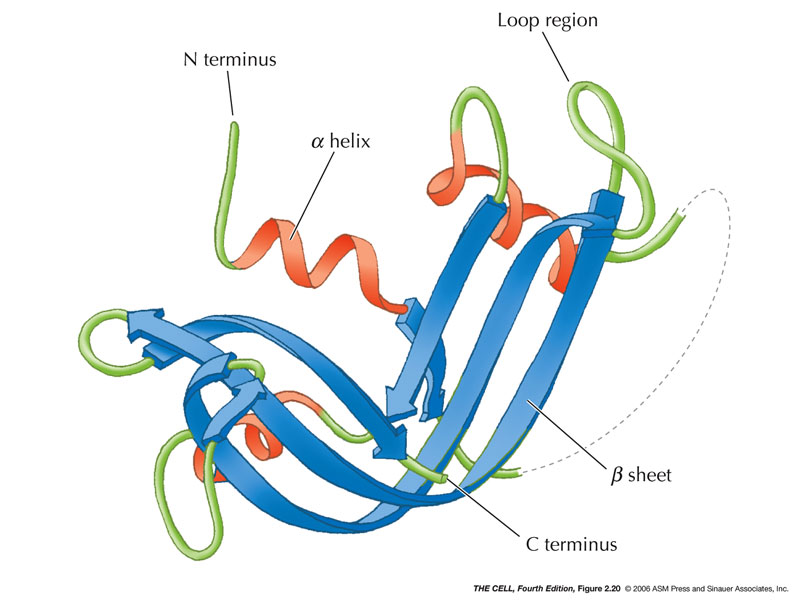
- Secondary Structure: This is the
local amino acid interactions that give 3D
structure.
- Alpha-Helix:
Hydrogen bonds form between amino acids holding it
in a right-handed helix and neutralizing the
polarity of the carboxyl and amino groups.
(Hydrophobic)(Pauling
video)
- Beta-Sheet:
Also held together by hydrogen bonds.
(Hydrophobic)
|
 |
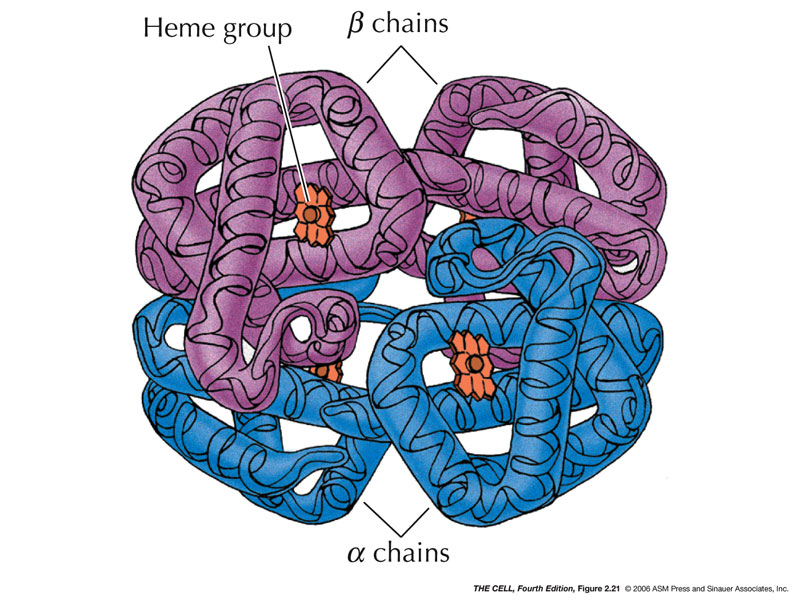
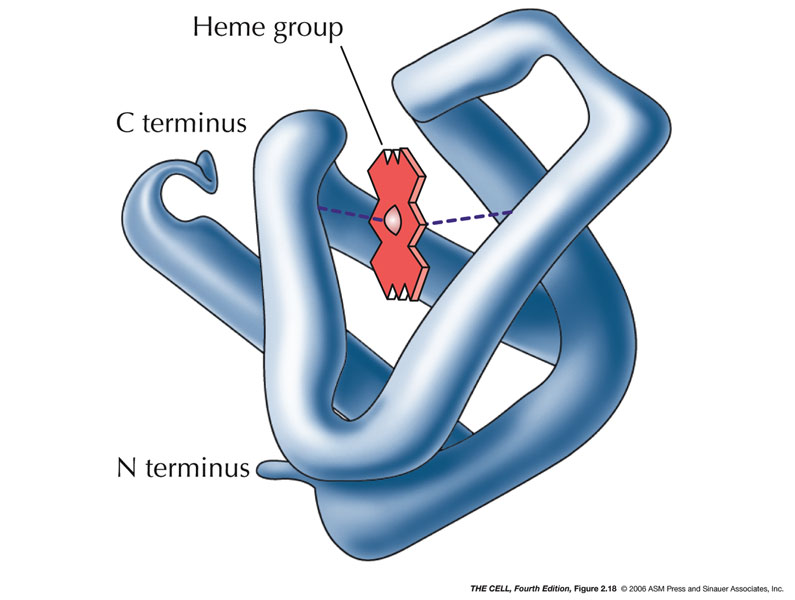
- Tertiary
Structure: Various
non-local
amino
acid interactions determine other 3D structures.
Proteins have structurally and functionally related
regions called domains.
|
 |
 |
|
|
|
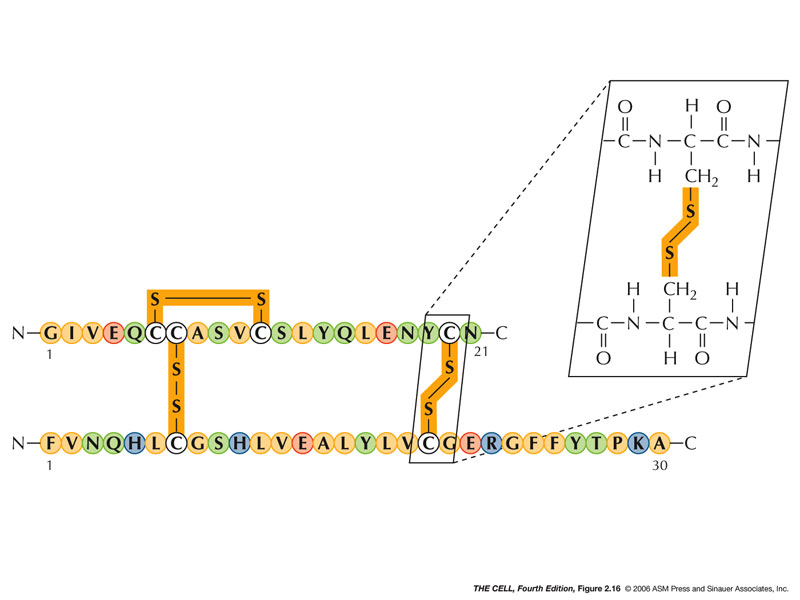
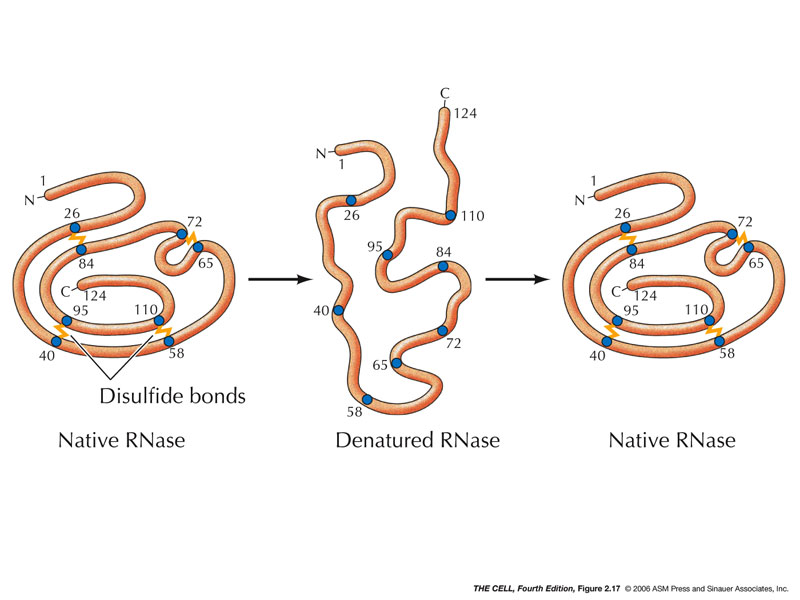
Disulfide Bridges
|
|
 Home
Home