http://upload.wikimedia.org/wikipedia/commons/thumb/8/87/Postnatal_genetics_en.svg/786px-Postnatal_genetics_en.svg.png
Gene Expression is Regulated: When
a gene is expressed, that means a specific polypeptide
(protein) encoded by that gene is being made. Not all
possible proteins are made all of the time or in all
tissues. For example, the globin genes of human hemoglobin
are differentially expressed. Adult hemoglobin is made up of
two α globins and two β
globins plus 4 iron-containing heme molecules. However,
the β globin is replaced by similar β-like
globins,
the γ globin (α2γ2)
in the embryo. So, before birth, the expression of
the β gene is turned
off, then after birth it is switched on. Conversely, the
expression of the γ gene is turned on
before birth, then it is switched off. This is temporal
regulation of gene expression. That is, the expression of a
gene (in a given tissue) is being switched on or off over
time. Globin gene regulation also illustrates spatial
regulation of gene expression, which is called tissue
specific gene expression. For example, the expression of the γ
gene in liver is great (the gene is "on") at 12 weeks after
conception, but at that same time in a different tissue,
like the bone marrow or spleen, the expression of the γ
globin gene is nil (the gene is "off"). This illustrates
tissue-specific regulation of gene expression. Just how does
the regulation of gene expression occur?
|
- Transcriptional
Regulation: The main mechanism
of regulating gene expression is by transcriptional
gene regulation. That is, when the gene is "on" it is
being transcribed. Its mRNA is being made and therefore
the polypeptide encoded by that gene is being made. When
the gene is "off" the transcription of that DNA segment
is blocked. Since no mRNA is being made, that
polypeptide will not be made. Besides "on/off"
regulation, transcriptional control may also determine
how much of the protein is made by controlling the
amount of the mRNA made during transcription. (We will
cover this topic next.)
- Post-Transcriptional
Regulation: Even though transcriptional control
is the central mechanisms of gene regulation, it is not
the only mechanism. Events that occur after translation
may be regulated and affect the amount of protein made.
This is called post-transcriptional control and includes
RNA processing, translational control and
post-translational control. (We will cover this topic
after we finish transcriptional regulation.)
- Changes
in
Gene
Copy Number: In some exceptional cases, gene
expression is affected by changing the number of copies
of a DNA segment. That is, to make more of a gene
product, more copies of the gene is made. Or,
conversely, a gene is not expressed because it has been
lost from the genome. Again, this is NOT the normal
mechanism of gene expression, but it does occur.
- History:
In the 1890s, Weismann proposed the germplasm theory,
which stated that the cells destined to become gametes
were set aside early in development. This basic
concept is correct, but Weismann was incorrect in
assuming that when development occurs, the germplasm
retains all the genetic material but the
differentiated cells lose the genetic material they do
not need. This model of differentiation is basically
incorrect. Early cloning experiments, like that of
Steward in 1958, were performed to answer this
question of whether or not differentiation involves
gene loss. Since it is possible to clone an
entire organism from a single cell, Steward and others
demonstrated that the cell must not have lost any
genetic material in the course of differentiation.
- Gene
Loss: However, in some rare cases, genes ARE
actually lost during the life of an organism, thereby
affecting the expression of the genetic material.
- Chromatin
Diminution: In the roundworm Ascaris and its
relatives, during early development the somatic
cells lose segments of their chromosomes. (This is
different from splicing!) The germ line cells,
however, retain all of the genetic material. (So
Weismann wasn't 100% wrong.) The amount of the
genome lost in these worms varies among species from
25% to 85%.
- Chromosome Elimination: In many
sciarid flies, entire chromosomes (not just pieces
of chromosomes) are lost in the the somatic cell
line.
|
- The Strange Case of Paramecium and
Tetrahymena: These single-celled ciliates
have a very strange
life cycle, which includes the formation of
two nuclei: a macronucleus and a micronucleus. The
macronucleus is the "active" nucleus of the critter
(its genes are being expressed: transcription is
occurring here). The micronucleus is inactive
in the mature ciliate and just for reproductive
purposes. The macronucleus is both an example of
gene amplification (see below) and gene loss. During
the life cycle, the micronucleus (after meiosis,
swapping micronuclei, and nuclear fusion) divides
and gives rise to both types of nuclei. One event
that occurs during the formation of the macronucleus
is the loss of specific sequences of the DNA called
IESs (internal eliminated sequences). (Again, this
is NOT splicing!) Which genes are to be eliminated
is an RNA mediated process similar to RNA
interference (later). The post-meiotic micronucleus'
entire genome is transcribed. Previously, the old
macronucleus was entirely transcribed and by a
comparison of these two whole-genome transcripts,
the determination of which segments to discard is
made. In this way, 50,000 or more segments (IESs)
are removed in the formation of the macronucleus.
(Strange critter!)
|

 |
- Cancer Cells: Many chromosomal
changes occur in tumors, including chromosome loss.
This loss may make the cancer more "viable."
|
- Gene
"Gain": In some cases, the number of copies
of a gene is increased.
- Repetitive
vs. Single-Copy DNA: Some genes are
normally present in multiple copies. Therefore, with
more copies of the gene, more gene product can be
made (rRNA genes). Eukaryotic genomes are typically
composed of three types of DNA, as determined by
hybridization studies (Cot values).
|
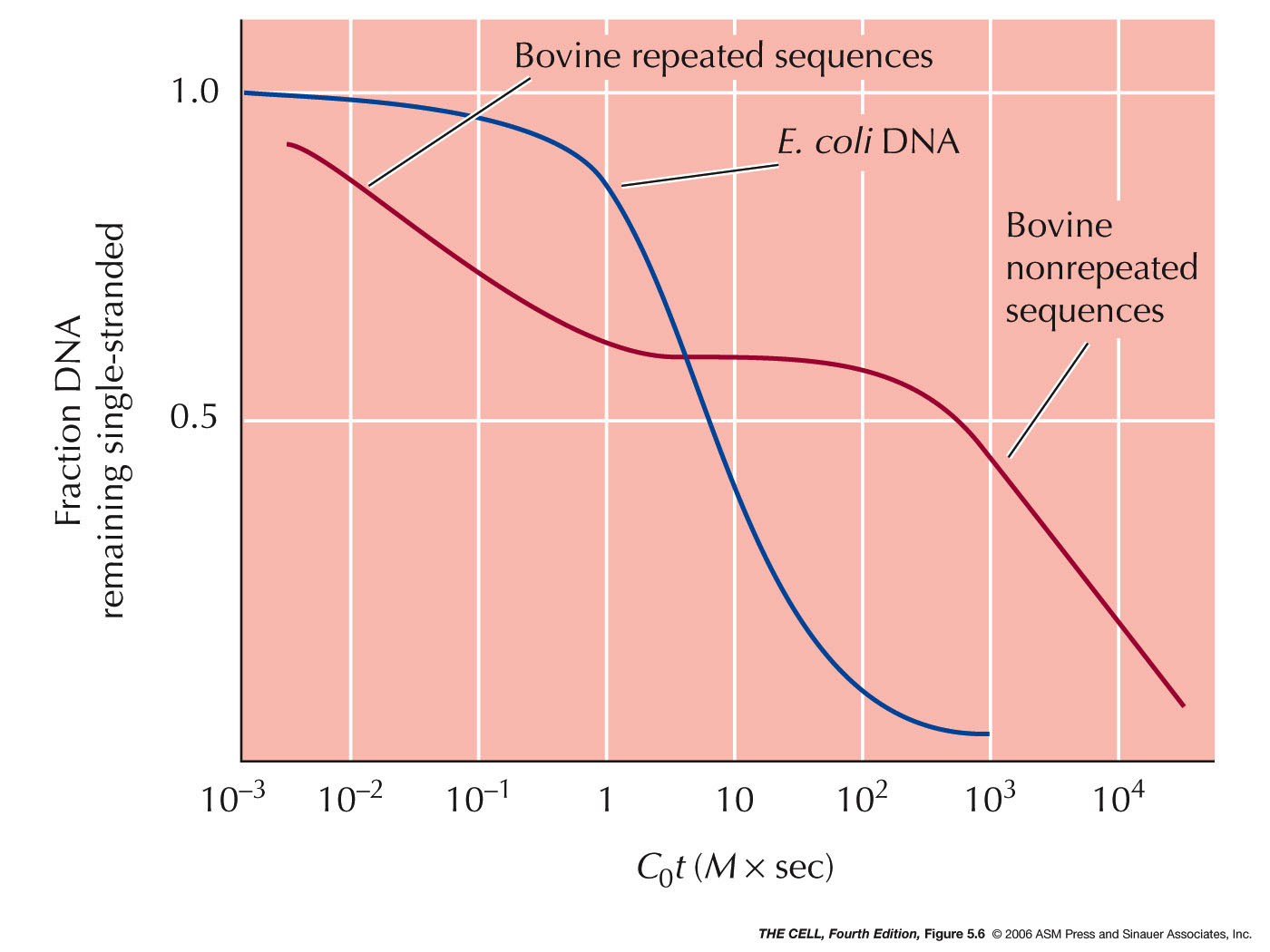
- Single-Copy (Unique-Sequence) DNA:
In humans, about 50% of the DNA is present only
once (one copy) in each genome (two copies per
cell). This includes gene coding for proteins.
This DNA hybridizes "slowly" (high Cot
values).
- Middle-Repetitive DNA: In humans,
about 40% of the DNA is moderately repeated, with
copy numbers between 10 and 1,000. This includes
the genes for rRNAs and tRNAs.
- Highly-Repetitive DNA: In humans,
about 6% or the DNA is present in very high copy
numbers as tandem repeats, with copy numbers of up
to 100,000. The repeated segment varies in length
from 5 to 300 nucleotides. This DNA is often
called satellite DNA (forms a satellite during
CsCl centrifugation). It includes transposable
elements and repeated segments unique to the
centromere and telomere. This DNA is usually
heterochromatin (heterochromatin is chromatin that
is condensed even during interphase--versus
euchromatin that decondenses during interphase and
includes active genes)(Dark
staining=heterochromatin, lightly
staining=euchromatin)
|
- Gene Amplification: In some cases,
DNA segments (or whole genomes) are replicated
without division.
- Amphibian Oocyte rRNA Genes: rRNA
genes in the giant amphibian oocyte are replicated
repeatedly, providing more templates to make this
ribosome component. This gene amplification can be
from 2000 to 1,000,000 fold.
- Drosophila
Polytene Chromosomes: Some dipteran
flies, including Drosophila, have giant
chromosomes in certain larval tissues. The Drosophila
salivary glands have these polytene
("many-stranded") chromosomes. Polytene
chromosomes have about 1000 chromatin strands and
show somatic pairing. They also have their
centromeres together in a structure called the
chromocenter.
|
- Ciliate Macronucleus: One other
event in the development of the macronucleus from
the micronucleus is the repeated replication of
the DNA (without cell division) producing a giant
nucleus with about 800 copies of the remaining
segments.
- Human Liver Cells: For some reason
(?), human differentiated liver cells often
undergo a round or two of replication without cell
division, making a polyploid cell (2-4 times the
normal number of chromosomes)(Polyploidy
in Liver Cells).
|
 Home
Home

