| DNA must be
able to carry out its basis role as the genetic material.
What must the genetic material be able to do? 1) carry
information, 2) copy that information (replication), 3) give
meaning to that information (determine traits). The
information is carried in the nucleotide sequence. We will
now look at the second of these roles. |
- The
Basic
Model of DNA Replication: Watson and Crick
proposed the basic
model
for
DNA replication at the time they proposed the
double helix: the template DNA specifies the nascent DNA
sequence by base pairing.
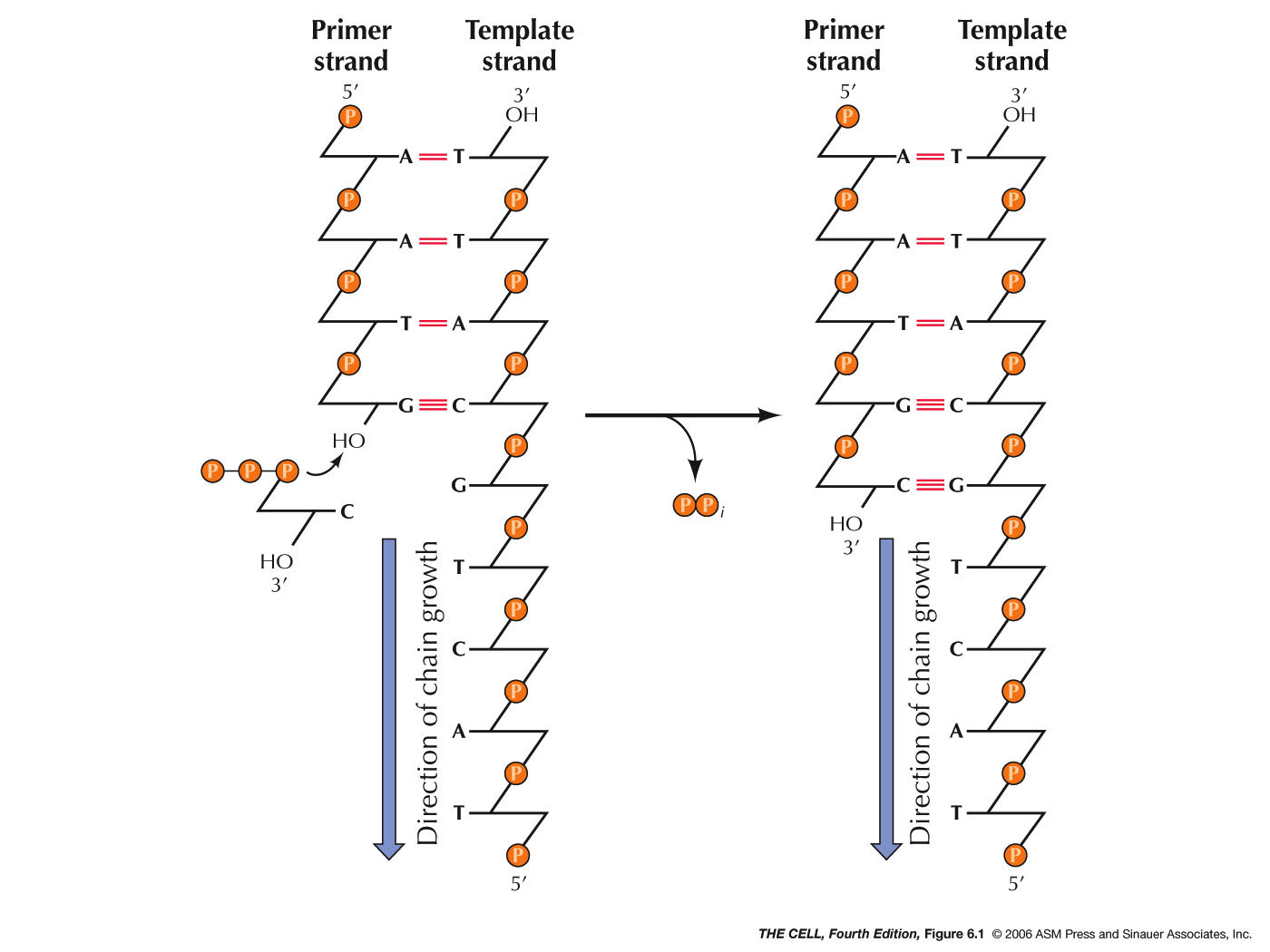
- DNA
Polymerase: In 1956, Kornberg discovered the
first DNA polymerase: E.
coli's DNA Polymerase I, not the main
replication enzyme (it works primarily in DNA repair).
DNA replication occurs at the replication fork. This
enzyme needs the 4 deoxynucleoside 5'-triphosphates,
primer DNA, and template DNA and directs the synthesis
of a DNA molecule following the sequence of the template
strand. When the nucleotide is added, nucleophilic attack
on the 3'-OH of the primer strand's last
deoxyribose cleaves off a pyrophosphate and a
phosphodiester bond is formed between the 3' O of the
primer and the 5' O of the newly added nucleotide.
The enzyme undergoes a 3D conformational change when
correct base pairing occurs, and then catalyzes the
formation of the phosphodiester bond. The enzyme
pyrophosphatase cleaves the pyrophosphate into two
inorganic phosphates, so the reaction cannot be
reversed. NOTE: DNA synthesis is a one way process. The
new DNA molecule grows in the 5' to 3' direction. That
is, it grow by primer extension at the 3' end. (Nucleotide nomenclature.)
|
|
|
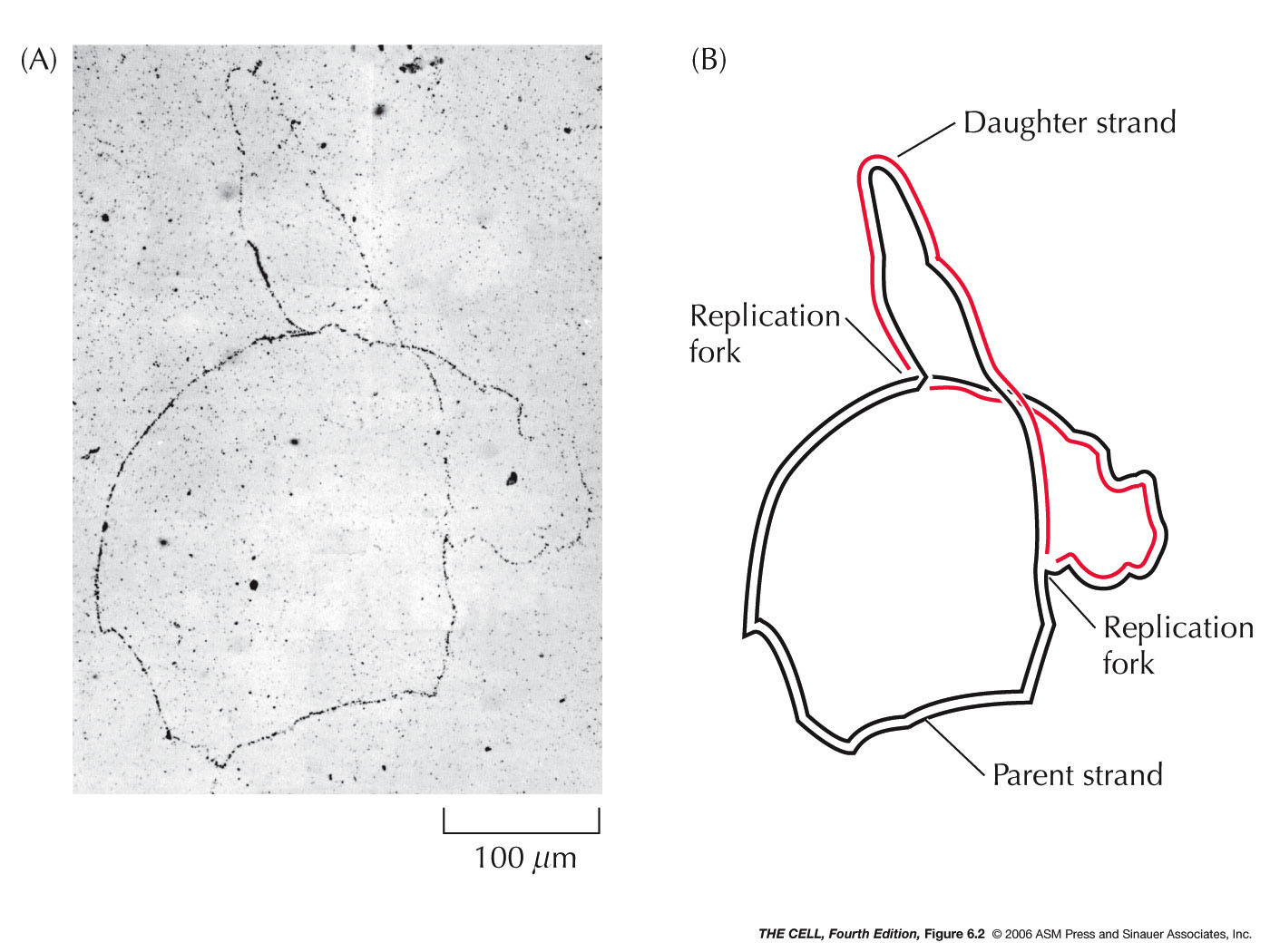
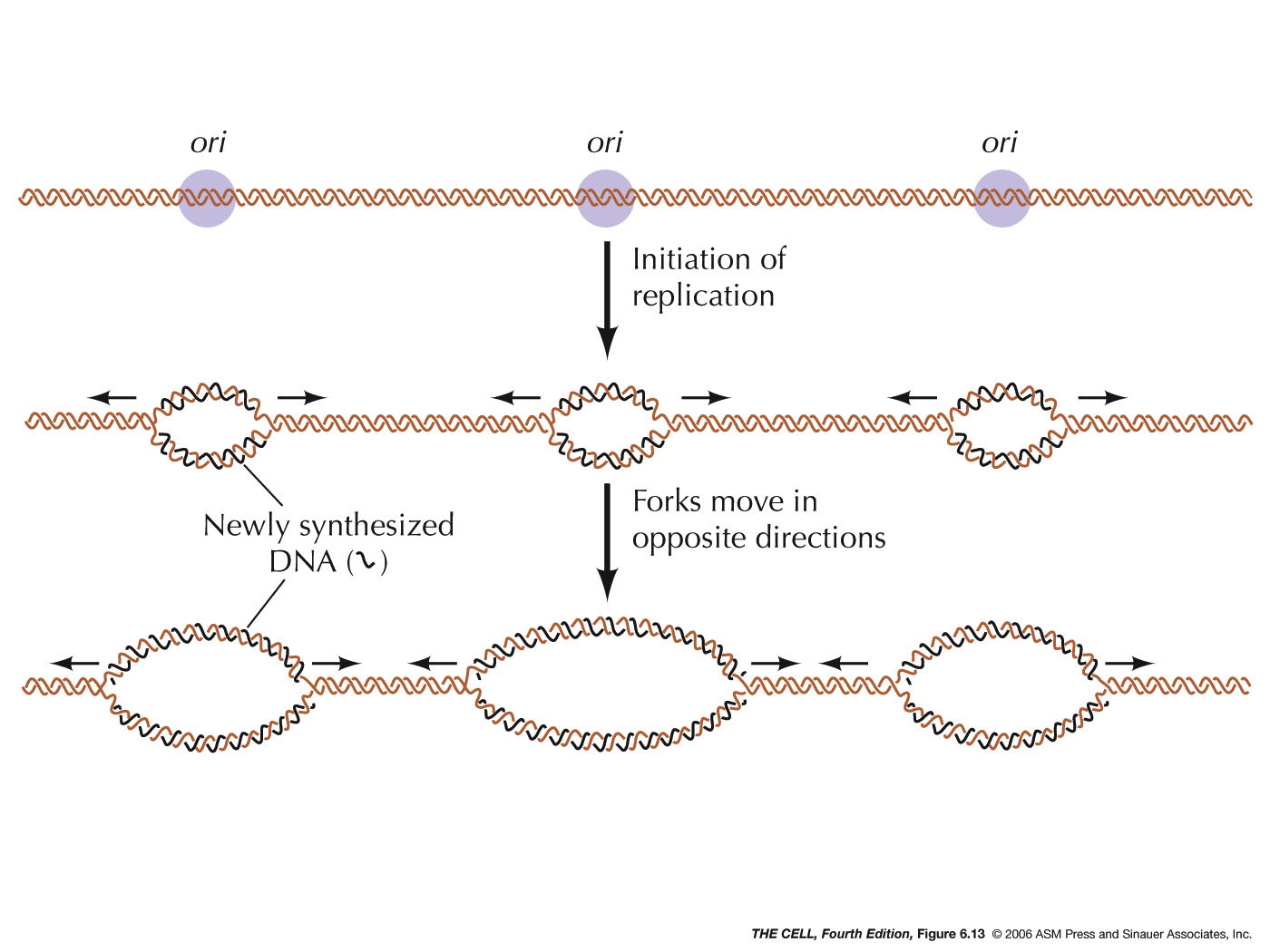
- Origin of Replication:
DNA replication begins at a sequence specific site. In
bacteria like E. coli, there is
one origin and replication proceeds
bidirectionally from that fork (autoradiography
photographs). In higher organisms, there are numerous replication
origins along each chromosome and replication is
bidirectional. Mammalian replication origins are
spaced 50 - 300 kb apart (kb=kilobase, 1000 bases). In
various tissues and developmental stages, the rate of
replication varies. The rate of replication is
apparently determined by how many replication forks are
activiated, not the speed of fork movement.
|
|
|
- Semiconservative DNA
Replication (pp. 110-111): Watson and Crick's
model of DNA replication can be called a
semiconservative model, since the newly made molecule
has one old strand and one newly made strand. Meselson and Stahl (original
paper) proved that DNA replication is
semiconservative in E.
coli in an experiment using DNA labeled with a
heavy isotope of nitrogen (15N
versus the normal 14N).
Bacteria were grown in growth medium with heavy
nitrogen. When these cells were transferred to medium
with light nitrogen and allowed to go through one round
of replication, their DNA was shown by CsCl density
gradient centrifugation to be hybrid in density (between
the density of all heavy and all light DNA). These
results were consistent with the semiconservative (but
not the conservative) model of DNA replication. Further
experiments also showed replication was not dispersive.
Later, Taylor, Woods, and Hughes showed DNA was
semiconservative in eukaryotes (plants) using
autoradiography.
|

|
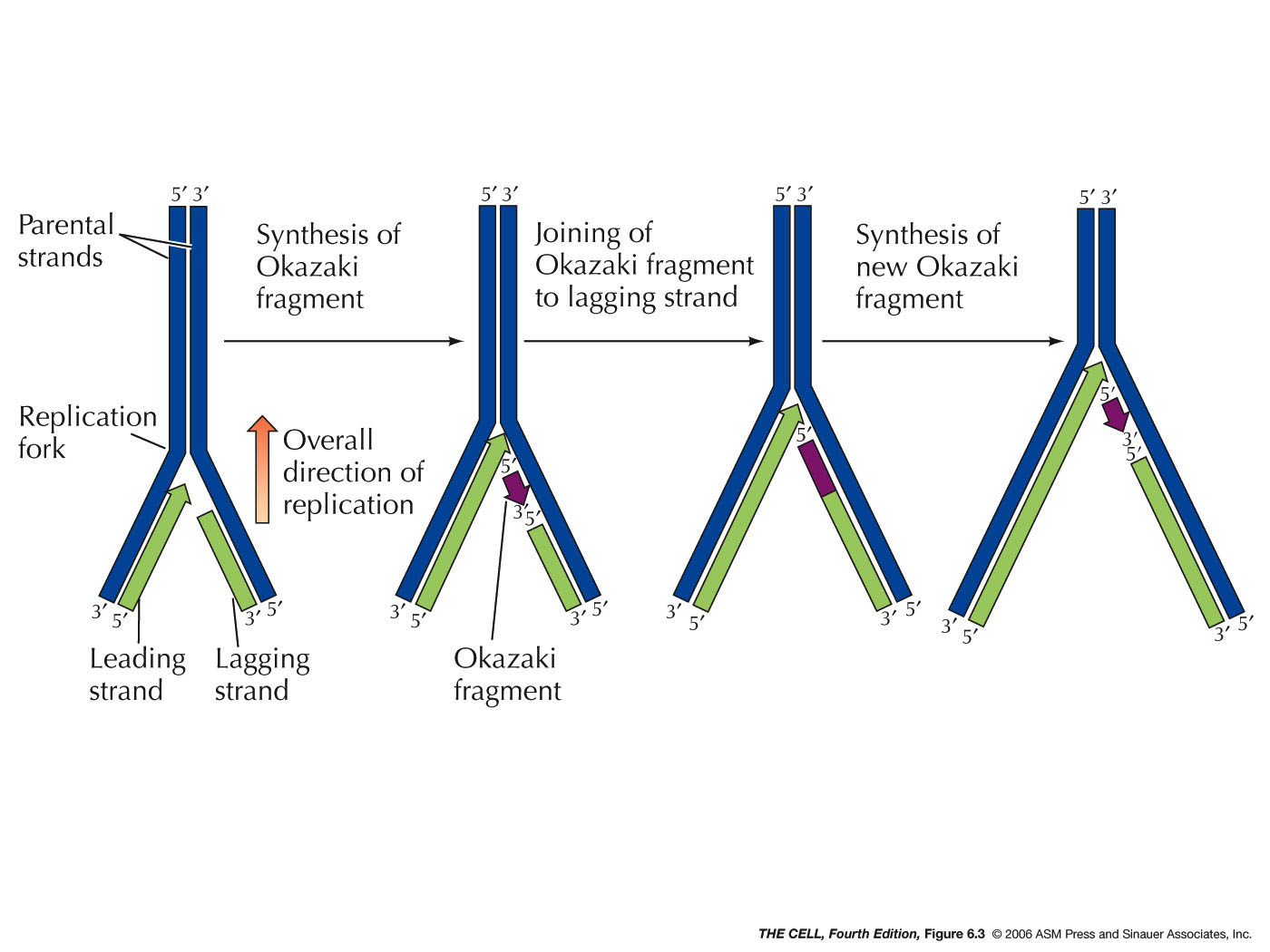
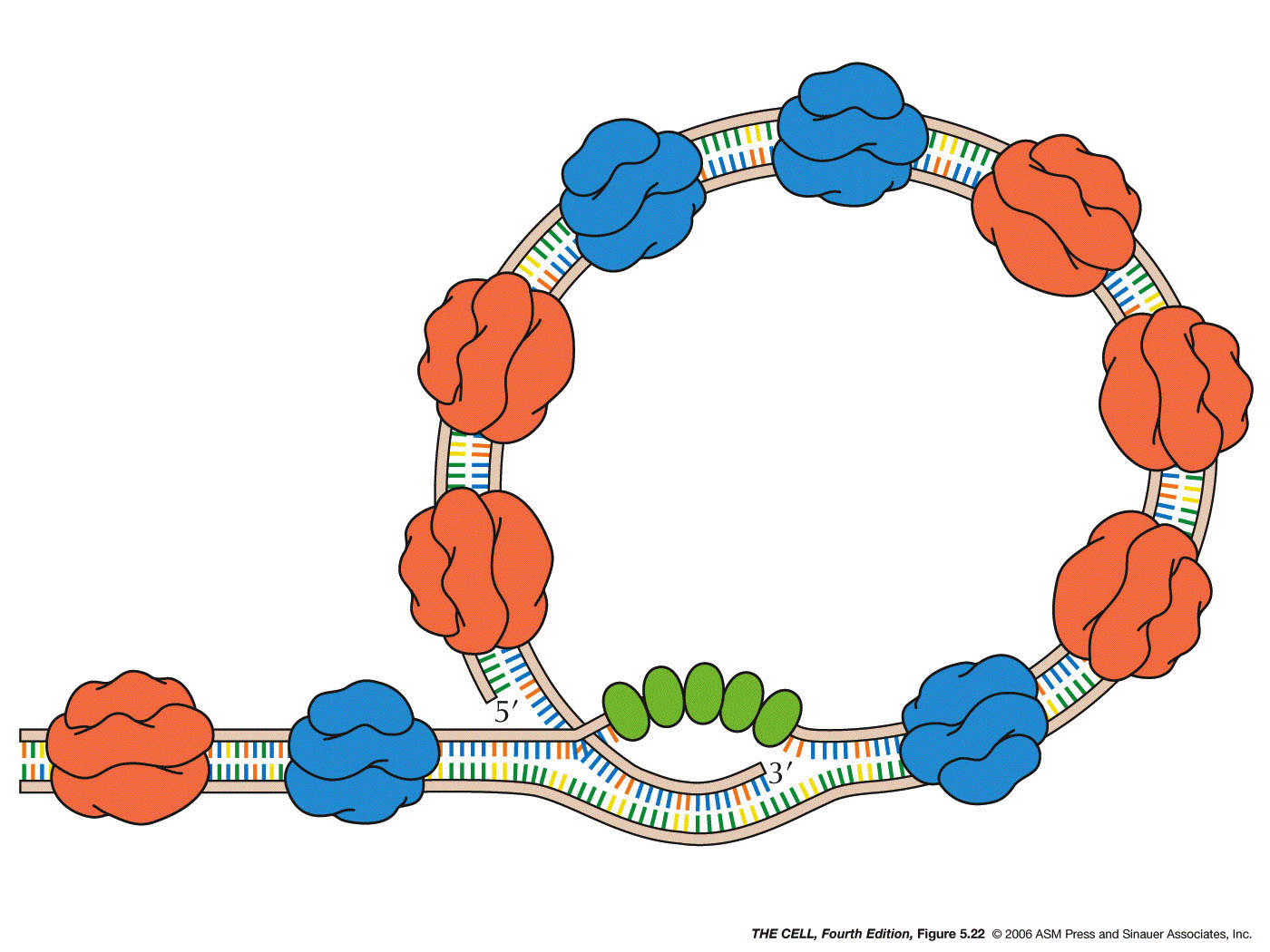
- Continuous
versus
Discontinuous
DNA Replication (Leading Strand versus Lagging Strand
Replication), An Overview: The problem of how
replication can occur on both strands since the process
only goes in one direction was solved by Okazaki with
the discovery of Okazaki
fragments. In E. coli, these are 1 - 3 kb
long fragments synthesized on the lagging strand. In
eukaryotes they are only 100 - 200 bp long. Leading
strand replication is continuous and does not involve
Okazaki fragments. Lagging strand replication is
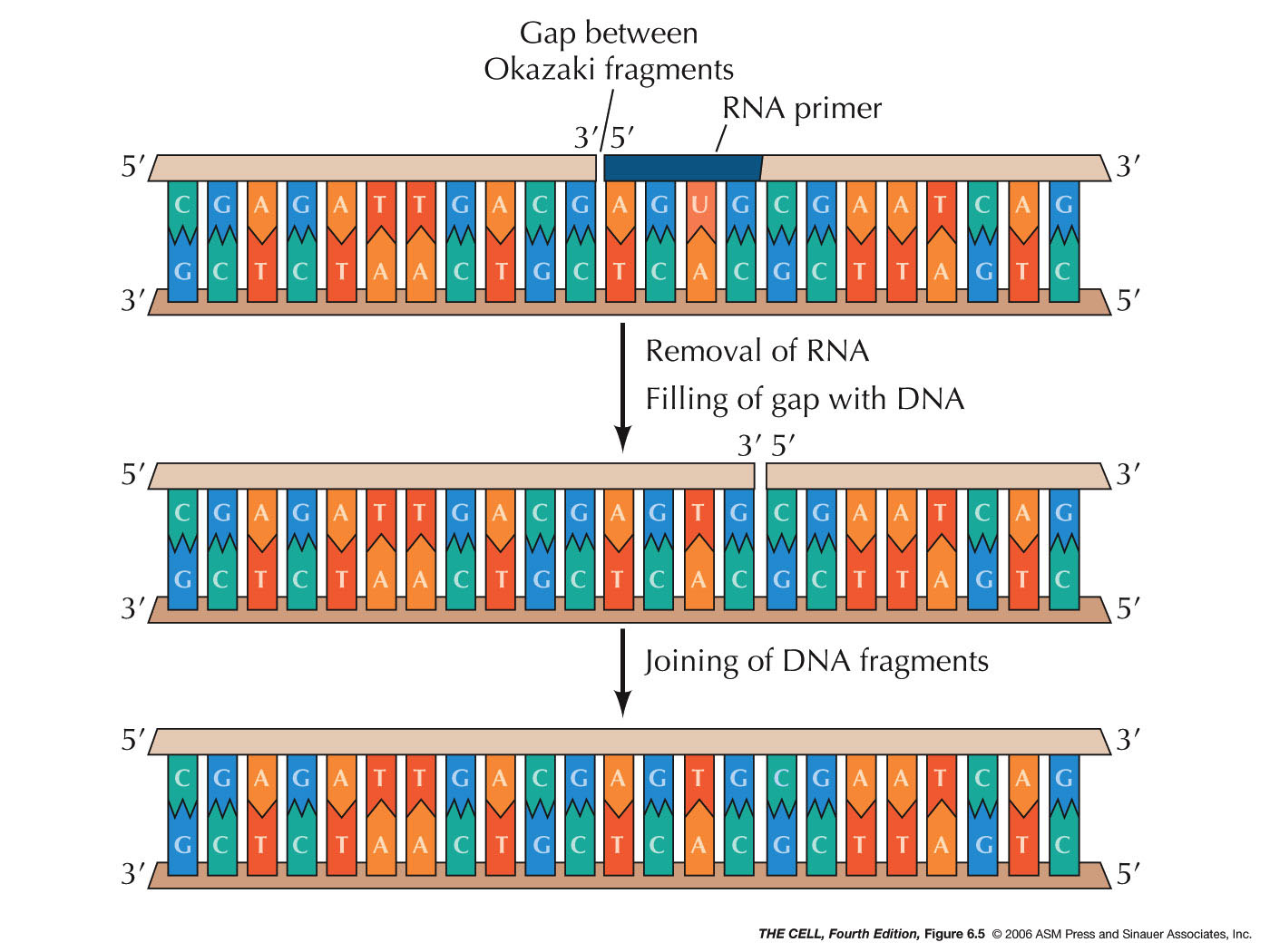
discontinuous and involves Okazaki fragments. Each
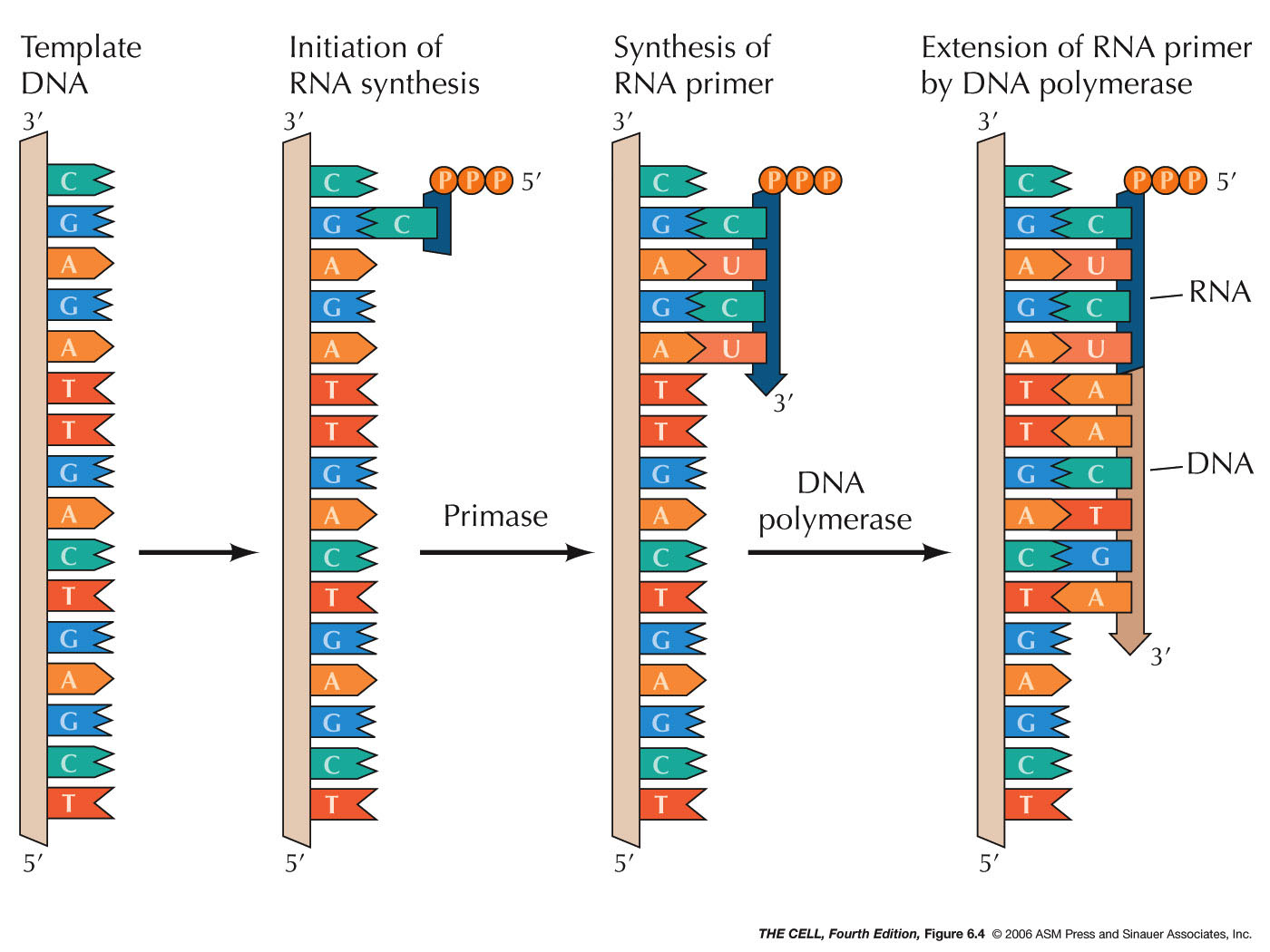
Okazaki fragment must starts with the synthesis of short
3-10 nt RNA primer
by action of the RNA polymerase called primase. These
RNA primers will be removed
by exonuclease activity and replaced by DNA. Finally,
when the growing 3' end of one Okazaki fragment reaches
the 5' end of the previous fragment, the resulting nick
is sealed by the enzyme DNA ligase. (DNA replication is
sometimes said to be semidiscontinuous.)
|
|
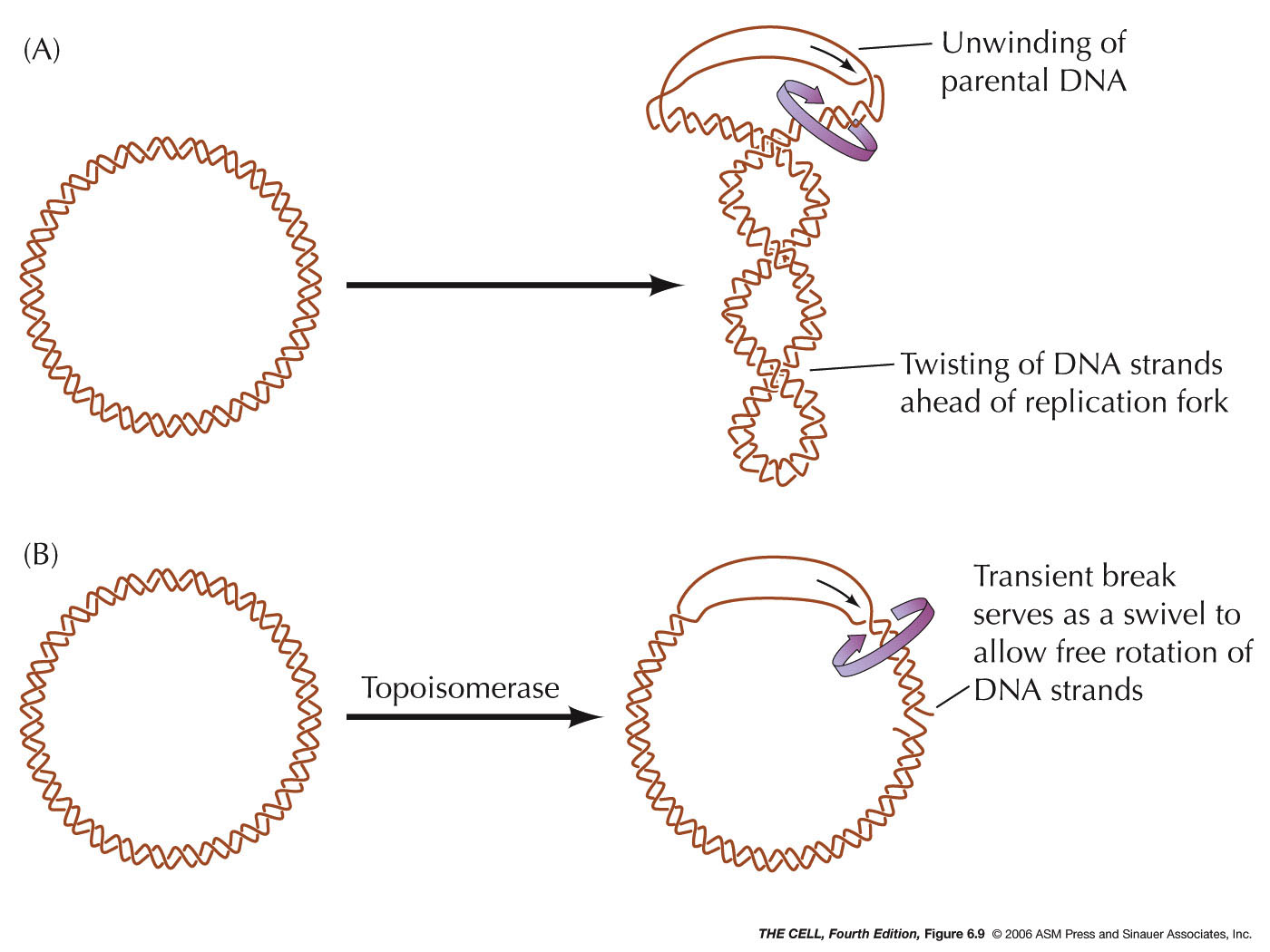
- Unraveling
the DNA: The action of topoisomerases
induce breaks in the DNA ahead of the replication fork
to allow for unwinding (swiveling). Type I
topoisomerases make single-strand breaks and type II
make double-strand breaks. Type II enzymes, like the
bacterial gyrase, are needed in replication-related
unraveling. These enzymes are also important in
untangling DNA molecules during DNA replication and cell
division and during crossing over. (Topoisomerase
info.)(Topoisomerase
I
and II video)
|
|
- The
Enzymes
of Replication: Many enzymes besides DNA
polymerase are required to replicate a DNA
molecule in vivo.
(Video)(more)
- The
DNA
Polymerases: This class of enzymes
synthesizes the new DNA, adding a nucleotide to the 3'
OH of a primer.
- Prokaryotic DNA Polymerases: There
are three DNA polymerases in E. coli: DNA
polymerases I, II, and III. DNA polymerase III is
the main enzyme of DNA replication, although I is
also involved (see below). The main job of DNA
polymerases I and II in the process of DNA repair.
- Eukaryotic DNA
Polymerases: Eukaryotes have three
polymerases involved in nuclear DNA replication: DNA
polymerases α, δ, and ε. A fourth polymerase,
γ, is the DNA polymerase that replicates
mitochondrial DNA. DNA polymerase δ was formerly
thought to be the main replication enzyme, while α
is involved with primase. α appears to have
DNA-dependent RNA polymerase activity (primase-like
enzyme: see Primase below). Some
new evidence indicates that ε is the primary
DNA polymerase working on the leading stand while δ
is the primary enzyme on the lagging strand. A
more recent article suggests that while δ is
the enzyme responsible for lagging strand
replication, it may also be involved with ε in
leading strand replication. (So the picture still is
not clear.)
- Primase:
This is a DNA-dependent RNA polymerase, like the ones
we will see later that make mRNA. Primase synthesizes
the short RNA primer and does not need a primer
itself, so it can start a new Okazaki fragment. In
humans, DNA polymerase α is composed of 4 subunits,
two of which begin the primer by synthesizing RNA
complementary to the template DNA. However, DNA
polymerase α also has DNA polymerase activity and
extends the RNA primer by adding DNA nucleotides.
Then, it is thought that DNA polymerase δ or ε takes
over to synthesize most of the Okazaki fragment. (DNA
polymerase α does not have the 3' to 5' exonuclease
activity needed for proofreading.)
- 5'
--->
3' Exonuclease Activity: An enzyme must
remove the RNA primer. This is accomplished by some
enzyme that has 5' ---> 3' exonuclease*
activity. The exonucleases involved in removing the
RNA primer remove one nucleotide at at time from the
5' end.
- Prokaryotes:
In E. coli,
DNA polymerase I (which has both 5' ---> 3' and
3' ---> 5' exonuclease activity in addition
to its polymerase activity) removes the RNA primer
and simultaneously synthesizes new DNA to replace
it.
- Eukaryotes:
In eukaryotes, a special exonuclease called RNase H
appears to work along with other exonucleases to
remove the RNA primer in the 5' to 3' directions.
DNA polymerase δ then returns to
synthesize DNA where the RNA primer was.
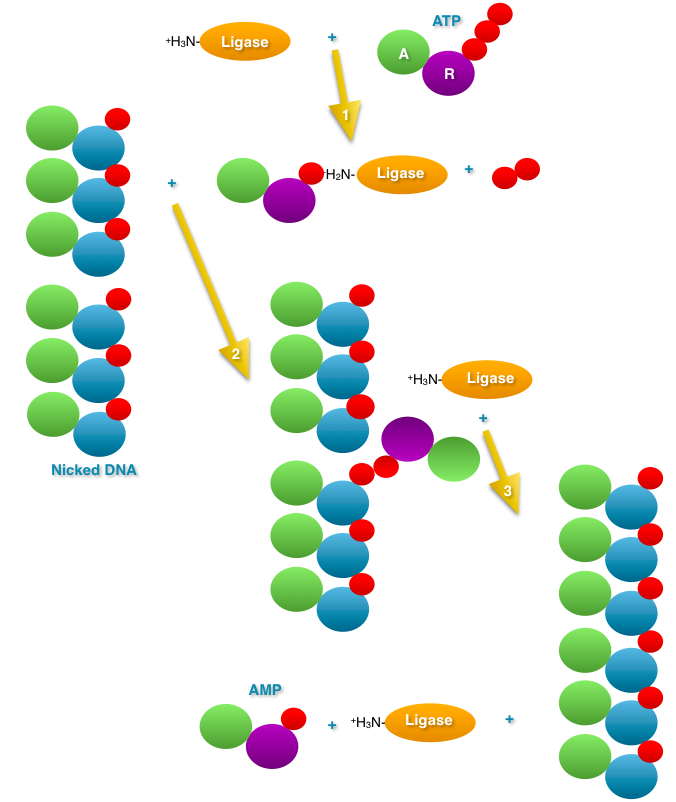
- DNA
Ligase: The nick left between two Okazaki
fragments must be sealed. DNA ligase joins the two
Okazaki fragments. Before ligation, there is a
phosphate on the 5' end of one fragment and a hydroxyl
on the 3' end of the other. DNA ligase first reacts
with ATP and AMP is covalently bond to ligase
(releasing pyrophosphatase). The AMP is then
transferred to the 5' phosphate of one fragment
(adenosine--phosphate--phosphate--then rest of the
primer 5' to 3'). Then, nucleophilic attack of the 3'
OH releases the AMP, ligating the two Okazaki
fragments. (Know the difference between a nick and a
gap and a double-strand break.)
 |
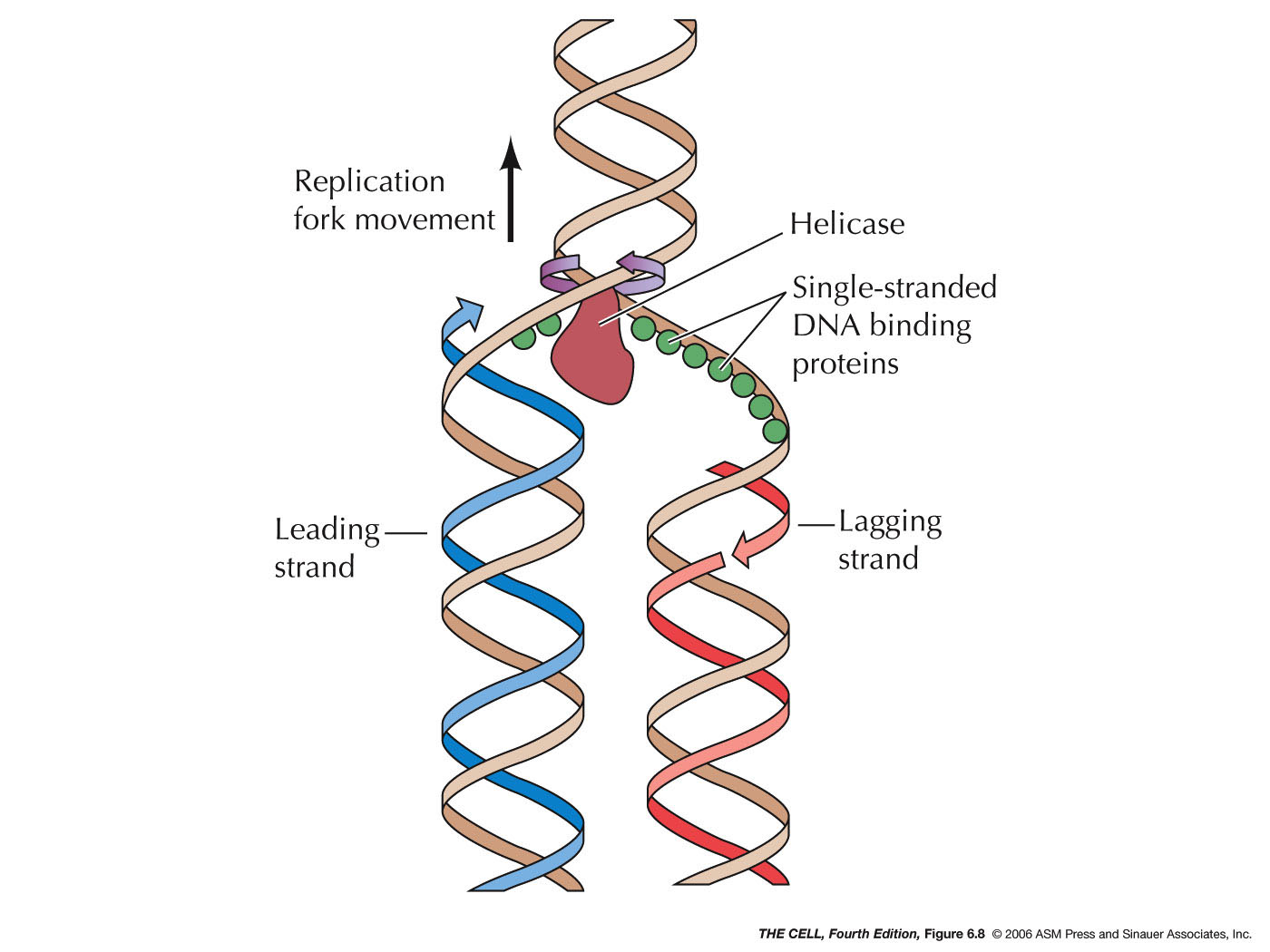
- Single-Stranded
DNA Binding Protein: These bind to and
prevent single-stranded DNA from collapsing on itself.
- Helicases:
This enzyme breaks the hydrogen bonds holding the two
strands together and unwind the two strands at the
replication fork.
|
|
- Clamp
Protein: Clamp-related proteins recruit and
hold DNA polymerase to the helix at the site of
polymerization. (Slides down the molecule)
- Topoisomerases:
Topoisomerases are a class of enzymes that
induce double-strand breaks or nicks in the DNA
backbone to allow for untangling. Type I
toposisomerases produce single-strand nicks while type
II topoisomerases (like DNA gyrase of E. coli) make
double-strand break. Type II are important in the
swiveling needed for prokaryote and eukaryote
untangling and unwinding during replication. Type II
can actually allow two helices to pass through one
another.
- A Video (from your text's
web site)
- DNA replication summary video
|
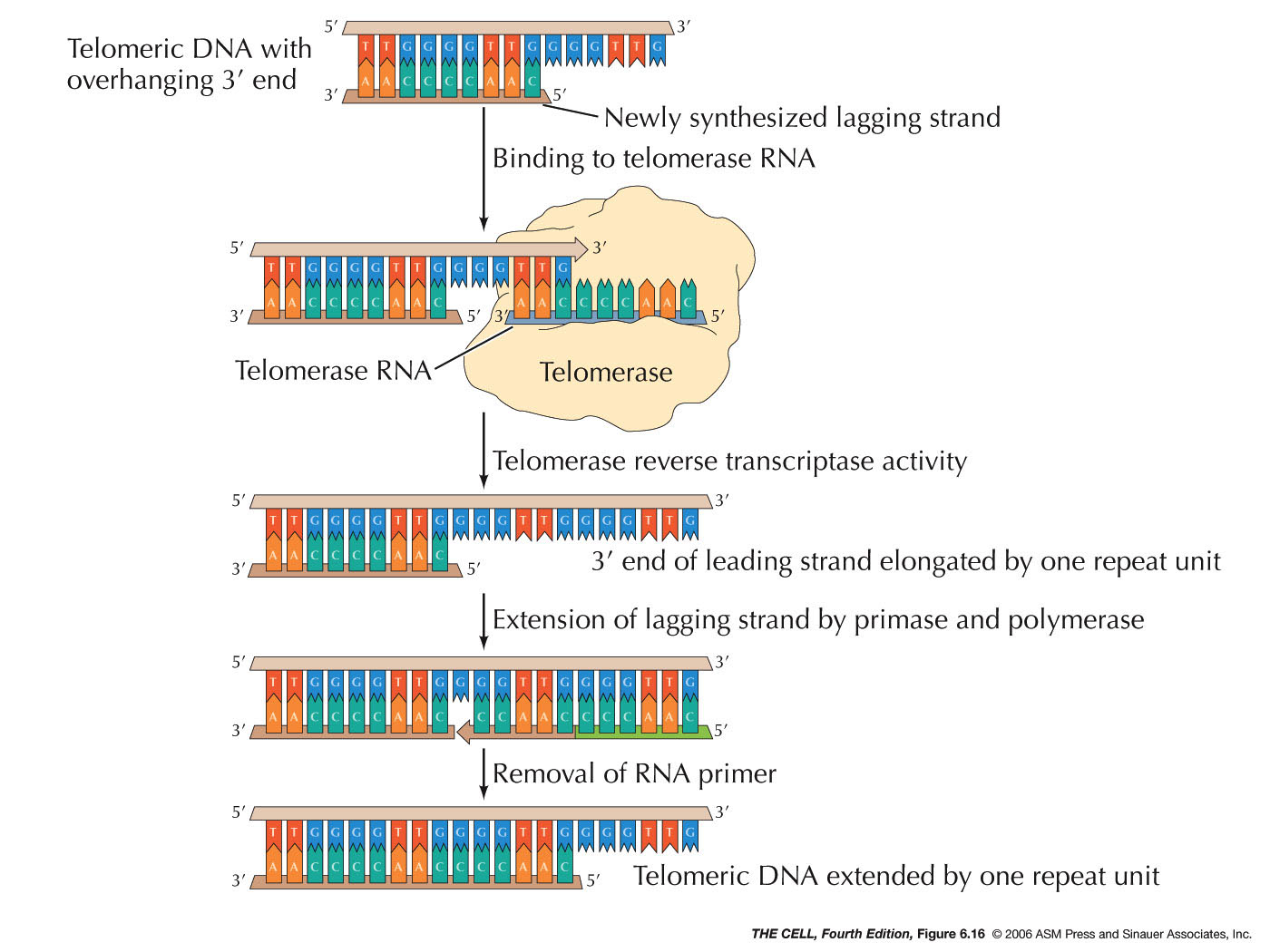
- The Replication of Telomeres
and Telomerase:
The ends of chromosomes are called telomeres. The
end of DNA molecules tend to react with other molecules
and cause problems. The loop structure of the telomere
helps get around this. Also, DNA replication at the
telomere occurs by way of the enzyme telomerase and
thereby gets around the 5' ---> 3' problem.
Telomerase is a reverse transcriptase that carries its
own template RNA (called TERC: telomerase RNA
component). TERC in humans is transcribed from an
RNA gene on the long arm of chromosome 3 (from a site
other than the telomere).
- Aging and telomeres:
Most human somatic cells lack much telomerase activity.
The length of the telomere correlates with the number
of divisions remaining before cell death. Cancer
cells have telomerase activity. (Some cervical cancers
have extra copies of TERC.)
- 2009
Nobel
Prize
(Medicine) to Telomere Researchers.
- Research
News:
Telomeres,
telomerase, and cancer.
*Nucleases are enzymes that digest nucleic
acids. Some are specific for DNA and are called DNases. Some
DNases digest only double stranded DNA and some digest only
single stranded DNA. Some nucleases are specific for RNA and
are called RNases. Some are non-specific and digest either
DNA or RNA. Nucleases are also classified according to
whether they break the nucleic acid backbone internally
(endonucleases) or cleave off one nucleotide at a time from
one end (exonucleases). |
 |
- DNA Replication
References:
|
 Home
Home










