| DNA's role as the genetic material
includes 1) carrying information (in its base sequence), 2)
copying that information (replication), and 3) giving
meaning to that information (determining traits). Genes
accomplish their job in this last role by directing the
activity of the cell, primarily by determining which
proteins (including the all-important enzymes) the cell
makes. By determining what enzymes the cell makes, the DNA
controls all of the complex chemical reactions that go on in
the cell (since enzymes control all of those reactions).
This process of DNA-directed protein synthesis occurs in two
stages: 1) transcription (messenger RNA synthesis: copying
the genetic information from DNA to RNA) and 2) translation
(polypeptide synthesis: using the genetic information in RNA
to make a specific chain of amino acids). Beginning today,
we will take a detailed look at transcription. Transcription
is the process of making all of the cell's RNA molecules,
not just those used in protein synthesis what are called the
messenger RNAs (mRNAs). We will also take up what happens to
those RNA molecules after they are made (RNA processing). Transcription overview video. |
- Transcription: Transcription
is
DNA-directed
RNA
synthesis (synthesis of RNA using DNA as the template).
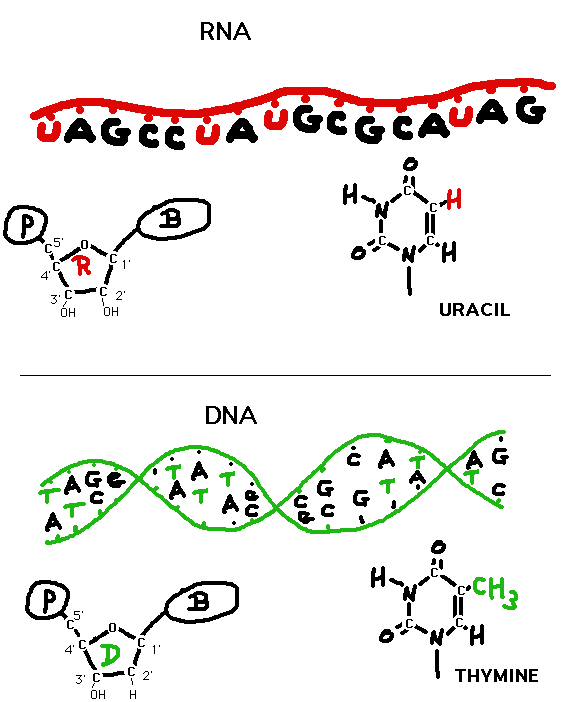
- In General: The
sequence
of a segment of a DNA molecule determines the sequence
of an RNA molecule. RNA is very
similar to DNA in structure, but 1) is usually
shorter, 2) is usually single stranded (but may form
double stranded loops call hairpin loops), 3) has
ribose in place of 2'-deoxyribose, and 4) has uracil
in place of thymine (uracil base pairs with adenine
just like thymine does). Transcription is the process
of making any RNA, whether it is mRNA, ribosomal RNA
(rRNA), transfer RNA (tRNA), as well as other small
RNAs (we will take up siRNAs and similar molecules
later). (Nucleotide
nomenclature)
- More Specifically: The
enzyme(s)
that
synthesizes RNA is called RNA polymerase. RNA
polymerase uses the nucleoside triphosphates (NTPs:
ATP, GTP, CTP, UTP) and RNA polymerization occurs just
like DNA polymerization does. That is, it begins with
the 5' end and the new RNA molecule grows in the 5' to
3' direction (a nucleotide is added to the 3'-OH). RNA
polymerase uses one strand of the DNA molecule as the
template strand with the 4 bases of DNA (A, G, C, T)
specifying which RNA nucleotides will be added (U is
added when the template DNA nitrogen base is A). As
with DNA replication, the newly-made RNA molecule is
antiparallel to the template DNA. However, unlike DNA
polymerase, RNA polymerase does not need a primer but
can add start a new RNA molecule with a single NTP
(therefore, the first nucleotide of an RNA molecule
has 3 phosphates on its 5' end). Only one strand of
the DNA double helix is used as a template. The
strand that is copied is called the template strand
and the strand not used is the non-template strand.
Any RNA molecule made by transcription is called a
transcript.
|
 |
- Transcription
in Prokaryotes: The details of transcription
were first elucidated in the bacterium E. coli.
|
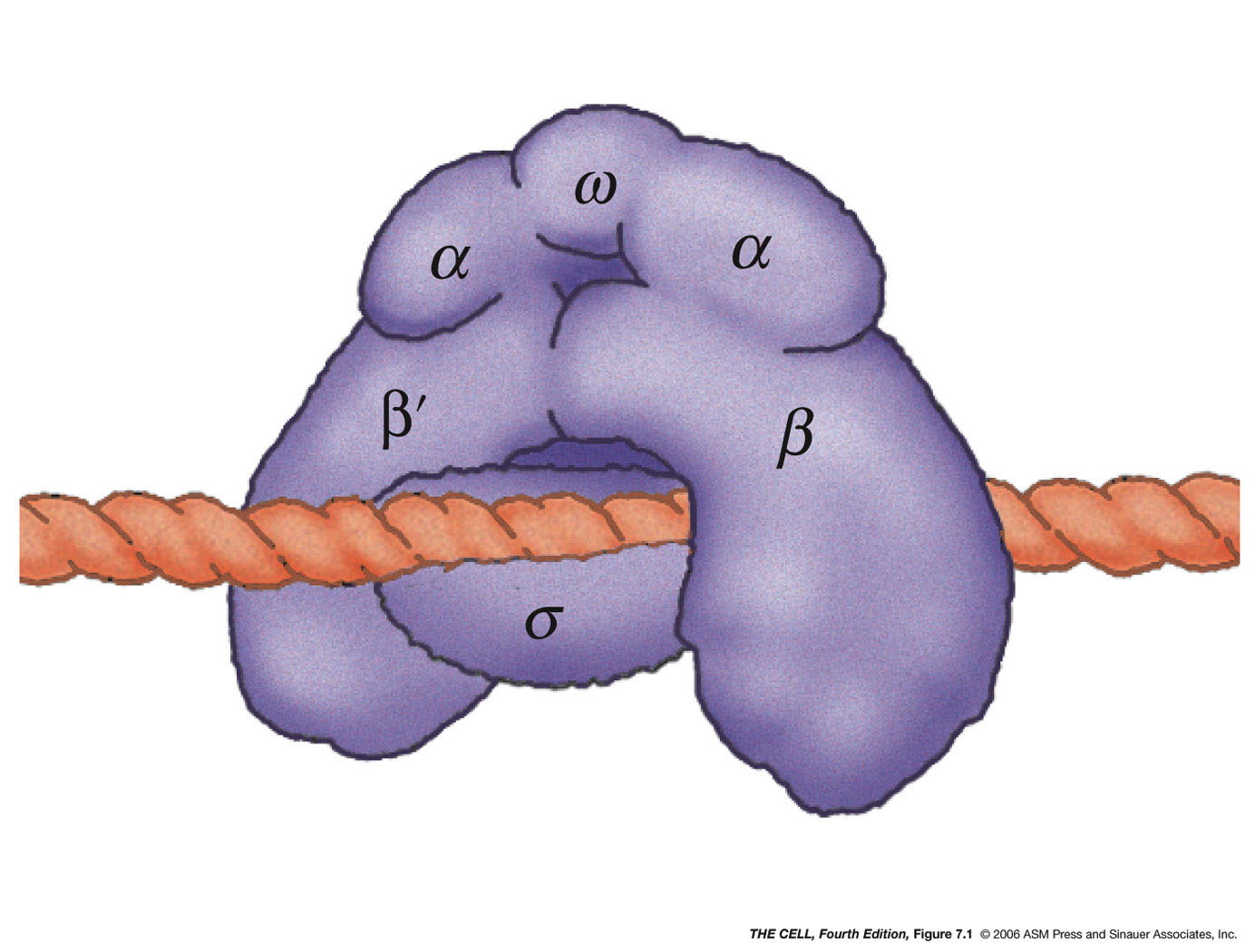
- E. coli's RNA
Polymerase and Its Action: E. coli
has only one RNA polymerase which makes all of the
cell's RNA. The enzyme consists of 6 subunits
(polypeptides)(α2,
β, β', ω, σ). This make up the holoenzyme.
However, the σ subunit is
loosely attached and the enzyme without σ
will still synthesize RNA, however not as
efficiently and it will not begin at the right
place. Therefore, σ is
needed for properly initiation of an RNA molecule
(needed to start at the right spot on the
DNA--called the promoter). This RNA polymerase
without σ is called the core
enzyme. RNA synthesis can be divided into three
phases: initiation, elongation, and termination.
|
 |
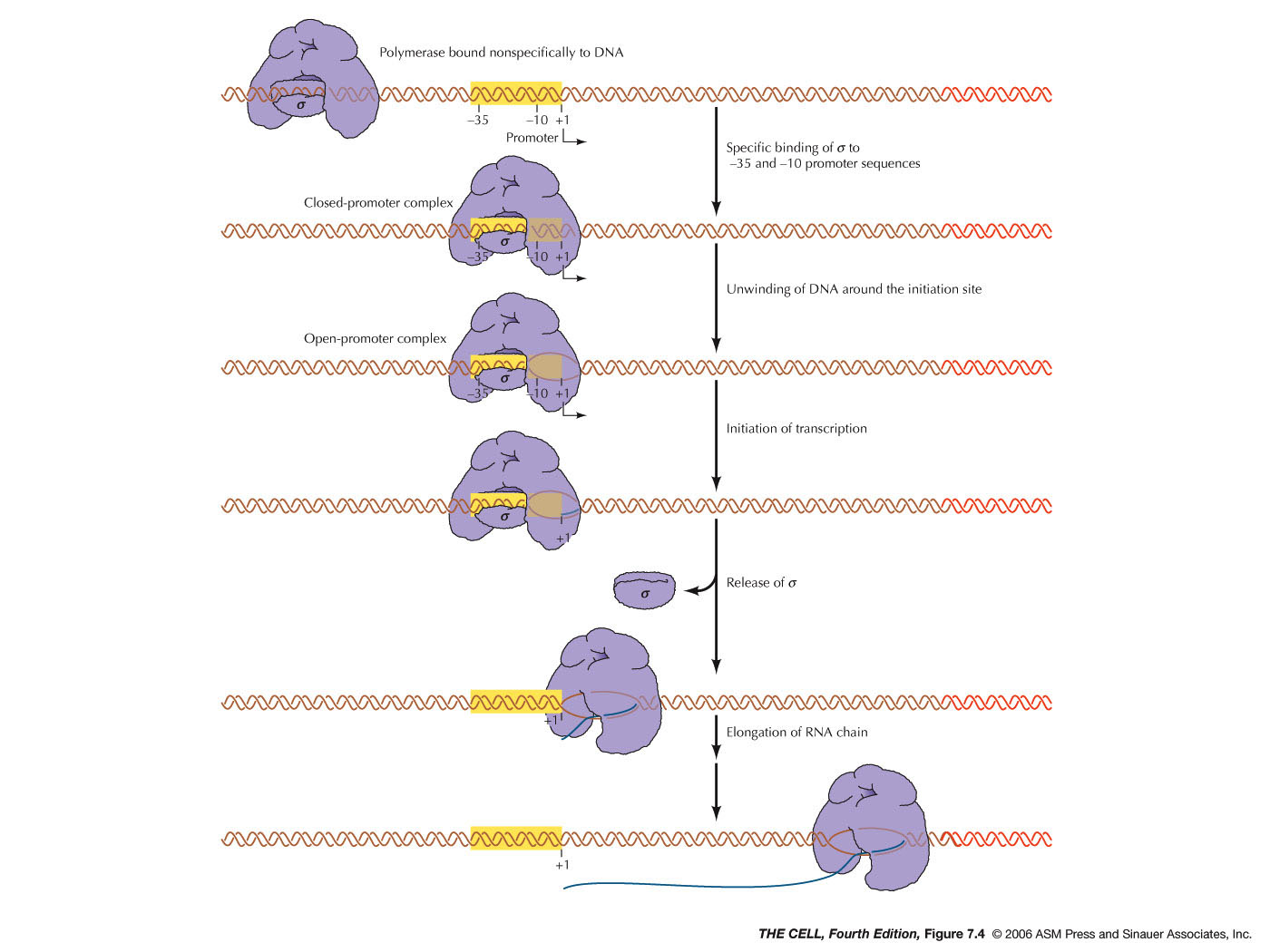
- Initiation:
The initiation
of mRNA synthesis begins at a site on the DNA
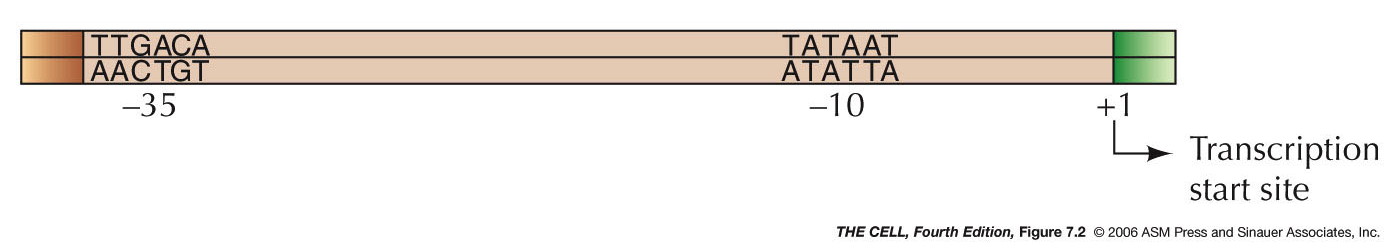
molecule called the promoter (RNA polymerase binds
here). The promoter
has two important regions (each about 6 nt long)
at -10 and -35 (this means 10 and 35 bp
upstream from--or before--the site where mRNA
synthesis begins). σ binds
to
these
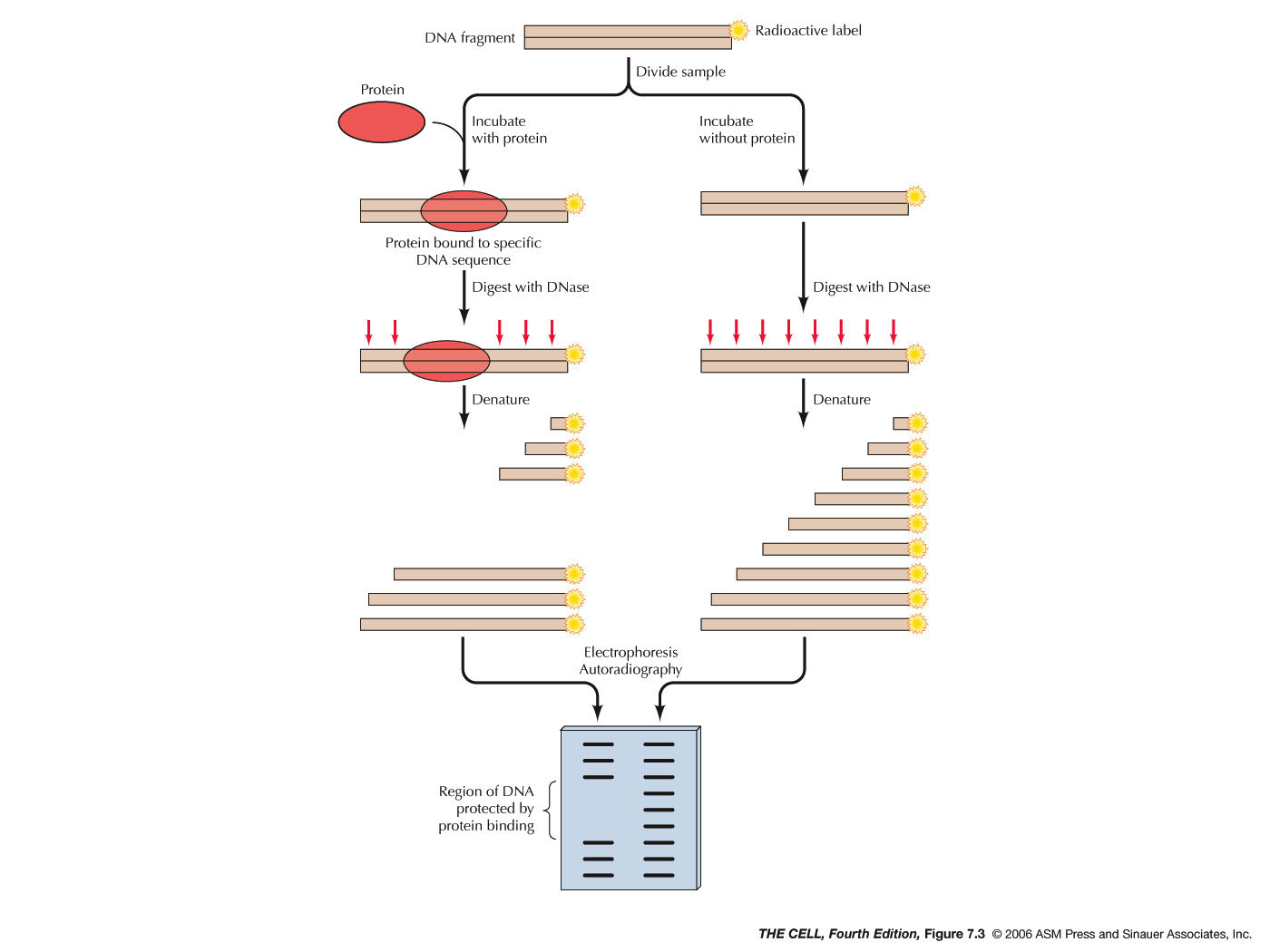
two sites. These sites were discovered by the
technique called protein footprinting
(read and know about footprinting in your text).
The entire RNA polymerase holoenzyme binds to a
sequence that spans the 60 bases from -40 to +20.
When the holoenzymes first binds to DNA, the DNA
is still closed (base paired). Upon initiation, a
12-14 bp segment opens up (about -12 to +2) and
RNA synthesis begins (the place where
transcription begins is designated +1).
- Elongation:
RNA synthesis continues as the RNA polymerase
"moves down" the DNA molecule. After about 10
nucleotides of RNA are synthesized, σ
is released (it is not needed after initiation and
can be used to start another transcript). As RNA
polymerase "moves" down the DNA molecule, it
"opens" the double helix ahead and closes it
behind, always maintaining about 15 bp open. In
the middle of this process, about 8-9 bases of the
3'-end of the mRNA are always base paired to the
template DNA strand.
- Termination:
When the mRNA molecule is completed, termination
releases the RNA from the DNA. This process occurs
by one of two methods.
|
 |
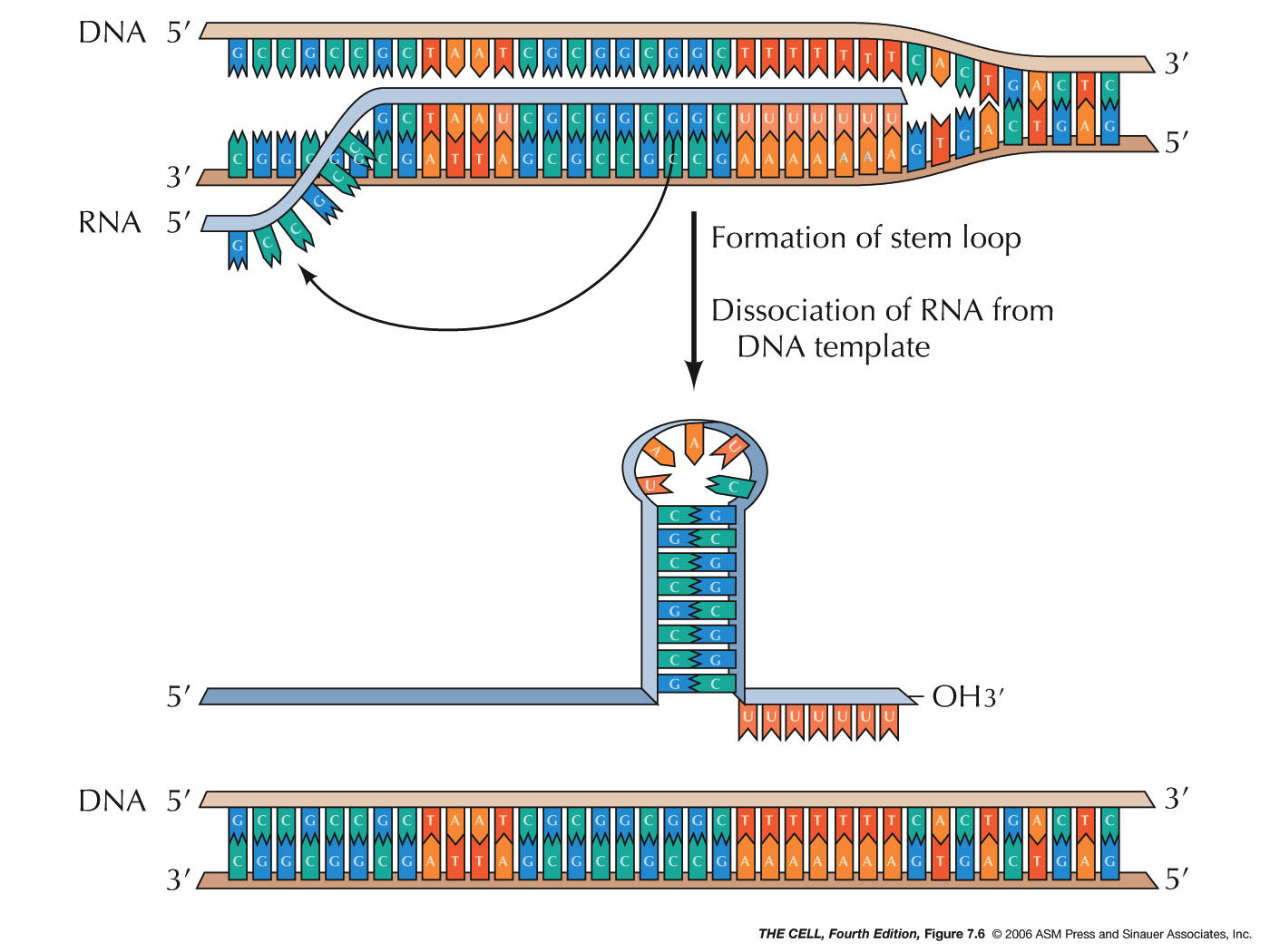
- GC-Rich
Sequence: In some cases, the end of the
gene is marked by a sequence rich in Gs and Cs
followed by a series of A bases (see figure).
When this GC-rich
region is transcribed, the RNA can base pair
with itself forming a hairpin loop, while
the RNA's U bases remain base paired with the
DNA's As. The stronger bonding of the internal
G-C pairs may disrupt the A-U pairs (A of
template DNA, U of RNA) causing the mRNA to be
released from the complex (this "pulls" the RNA
off of the DNA).
|
 |
- ρ-Mediated
Termination: In some cases, the ρ protein
binds
to extended mRNA and terminates transcription.
(Recent results concerning ρ-mediated
transcription termination is here. (Molecular
Biology, 5th ed., Weaver. McGraw-Hill
Publishers)
|
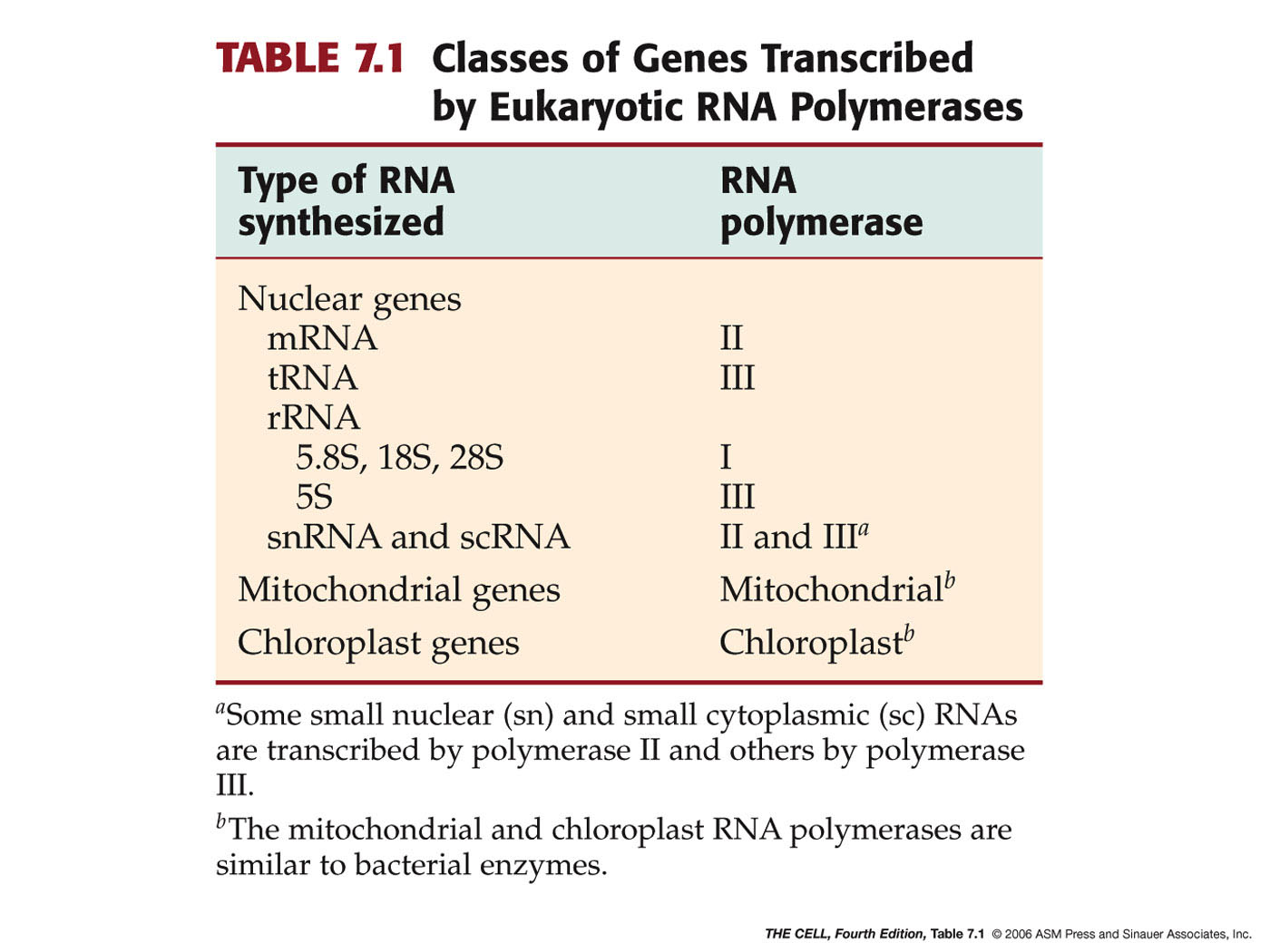
- Eukaryotes:
Eukaryotes have several
RNA
polymerases, each with specific tasks. They all
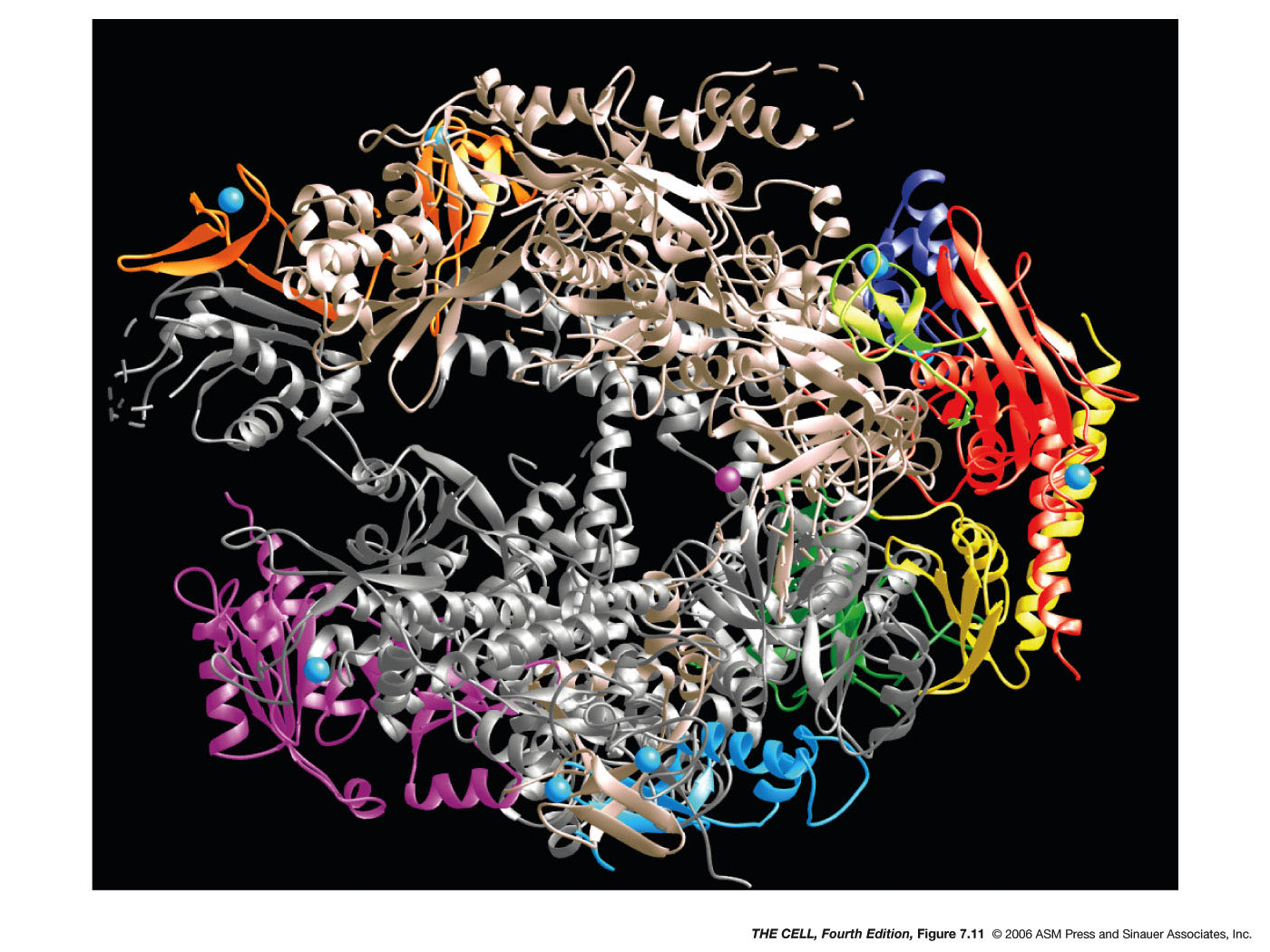
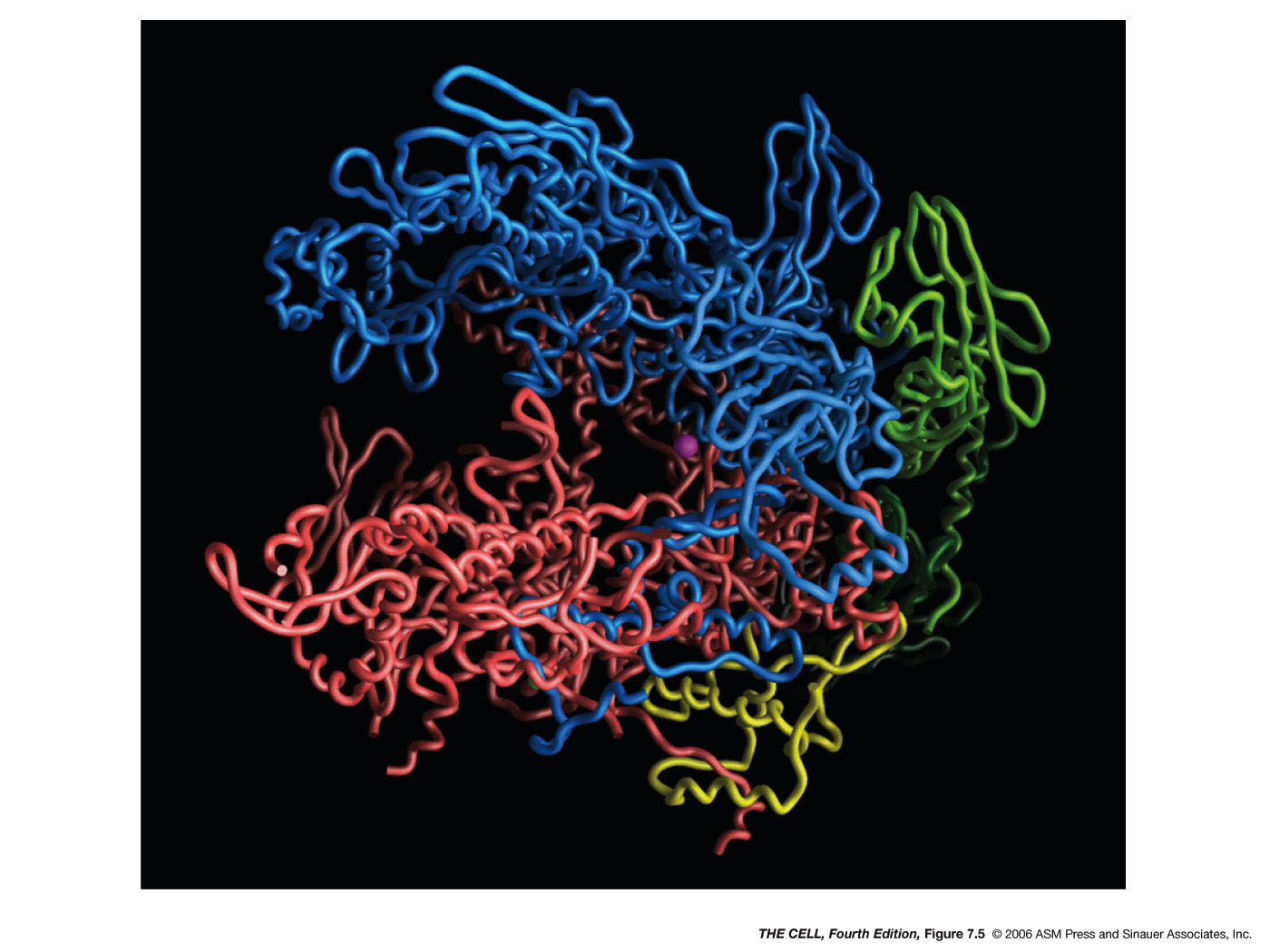
have 9 subunits, 5 of which are very similar to the E. coli
holoenzyme subunits. The 3-D structure of eukaryotic and E. coli RNA
polymerase is also very similar.
- RNA
Polymerase II: This enzyme makes
pre-messenger RNA. (We call it pre-mRNA because, as
we will see later, it must be processed to
become real mRNA.) So, this is the RNA polymerase
that transcribes the genes that make proteins. Only
RNA polymerase II has the CTD (C-terminus
domain--see diagram under Initiation below). (RNA
polymerase II
also makes some of the small RNAs we will see
later.)
  |
|
- Transcription
Factors: Eukaryotic RNA polymerase is
different from prokaryotic RNA polymerase because
it will not synthesize RNA without a number of
other proteins (factors) that are not in integral
part of the enzyme (unlike σ which
is
an integral part of E. coli RNA polymerase). These
factors for RNA polymerase II are of two types.
|
- General
Transcription
Factors: These factors are necessary
for transcription of any pre-mRNA to occur.
(Some will be cover in more detail in the next
topic.)
|
- Gene-Specific
Transcription
Factors: These factors are necessary
for the transcription of particular genes. We
will take these up later in this course when we
discuss the regulation of gene expression in
eukaryotes.
|
- Stages
of
Transcription: As in E. coli,
transcription involves initiation, elongation, and
termination.
|
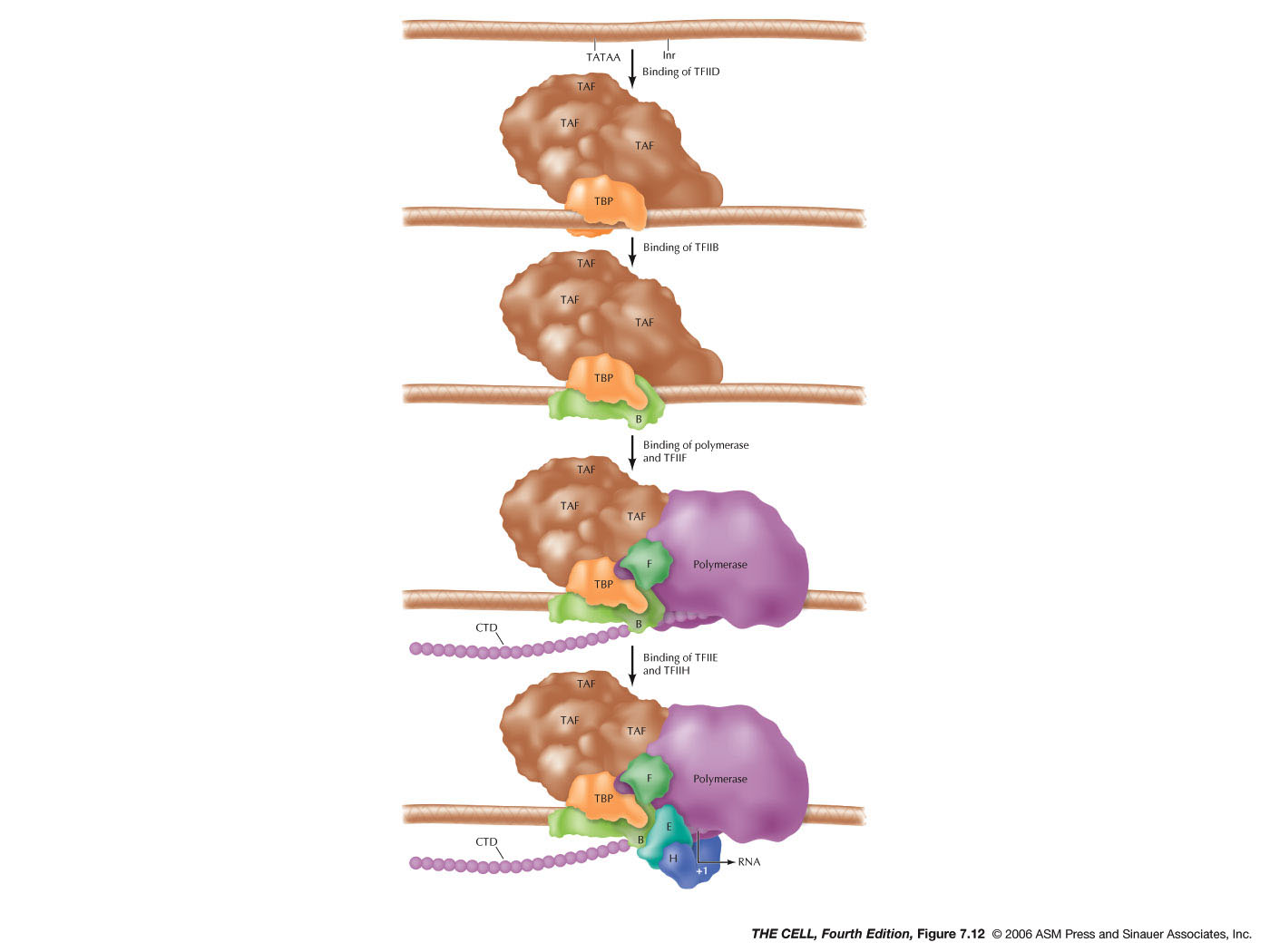
- Initiation:
The promoter usually includes a sequence called
the TATA box that is at about -30 to -25. (Some
genes do not have a TATA box but have other
sequences involved in initiation.) One of the
general transcription factors is TFIID which
binds to the promoter first and includes TBP
(TATA-binding protein)(see figure). Then TFIIB
binds which enables RNA polymerase binding
followed by the binding of other general
transcription factors. One of these is TFIIH
which includes the enzyme helicase, which
uncoils the DNA double helix.
|
 |
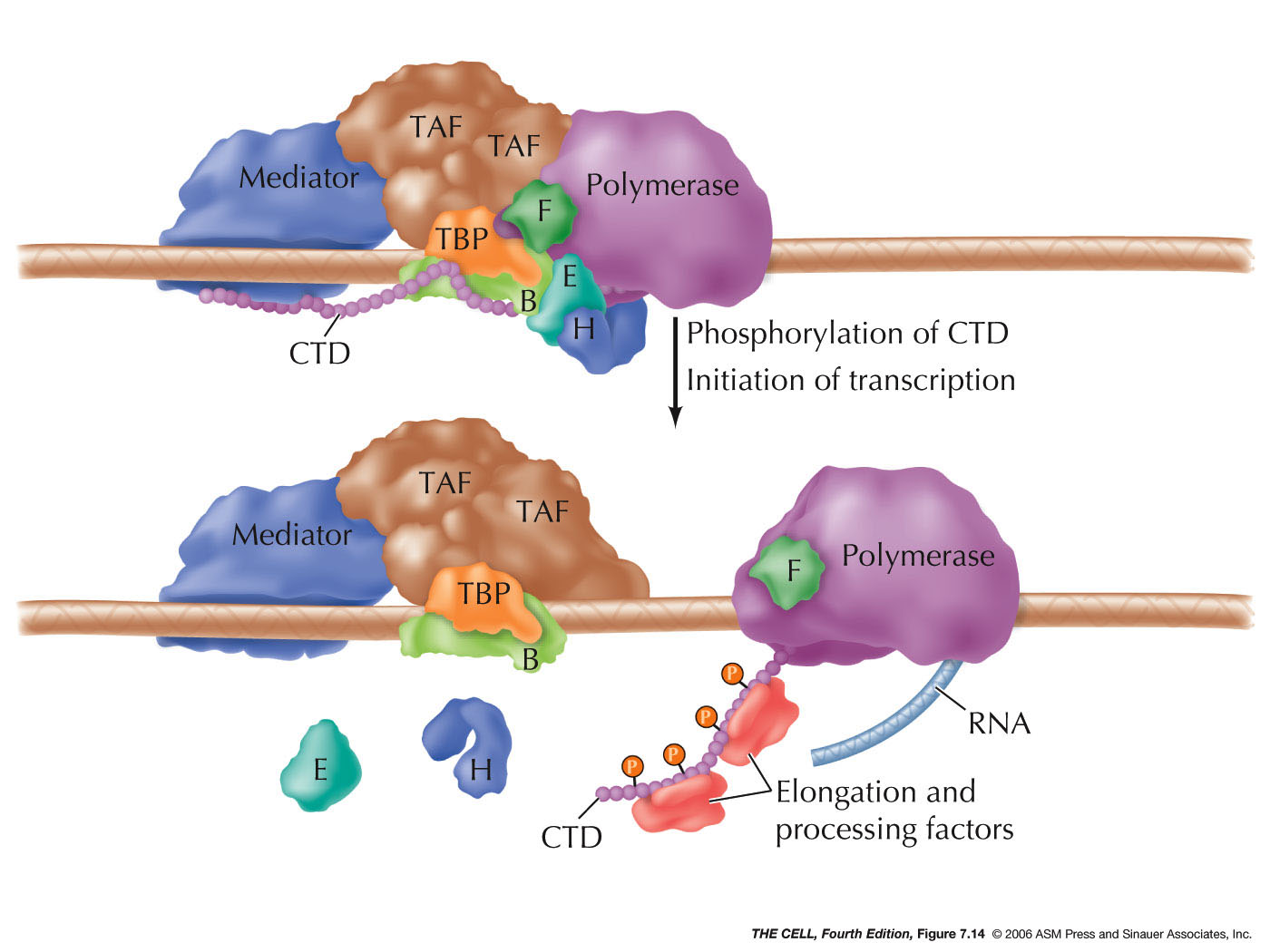
- Elongation:
Elongation of the pre-mRNA continues with the
aid of elongation factors.
|
 |
- Termination:
Termination involves cleavage of the pre-mRNA
and its polyadenylation, so it will be covered
under "RNA
Processing."
|
- RNA
Polymerase I and III: These RNA polymerases
transcribe short RNAs. RNA polymerase I makes the
larger rRNA molecules while RNA polymerase III makes
the tRNAs and the smallest rRNA and some other small
RNAs. (See "RNA
Processing" for more details on these two
enzymes.)
|
- Other
Polymerases: Chloroplasts and mitochondria
have their own RNA polymerases.
|
- RNA Processing:
When transcription is finished, the newly made RNA
molecules are altered, in some cases considerably. (Recent
information!)
|
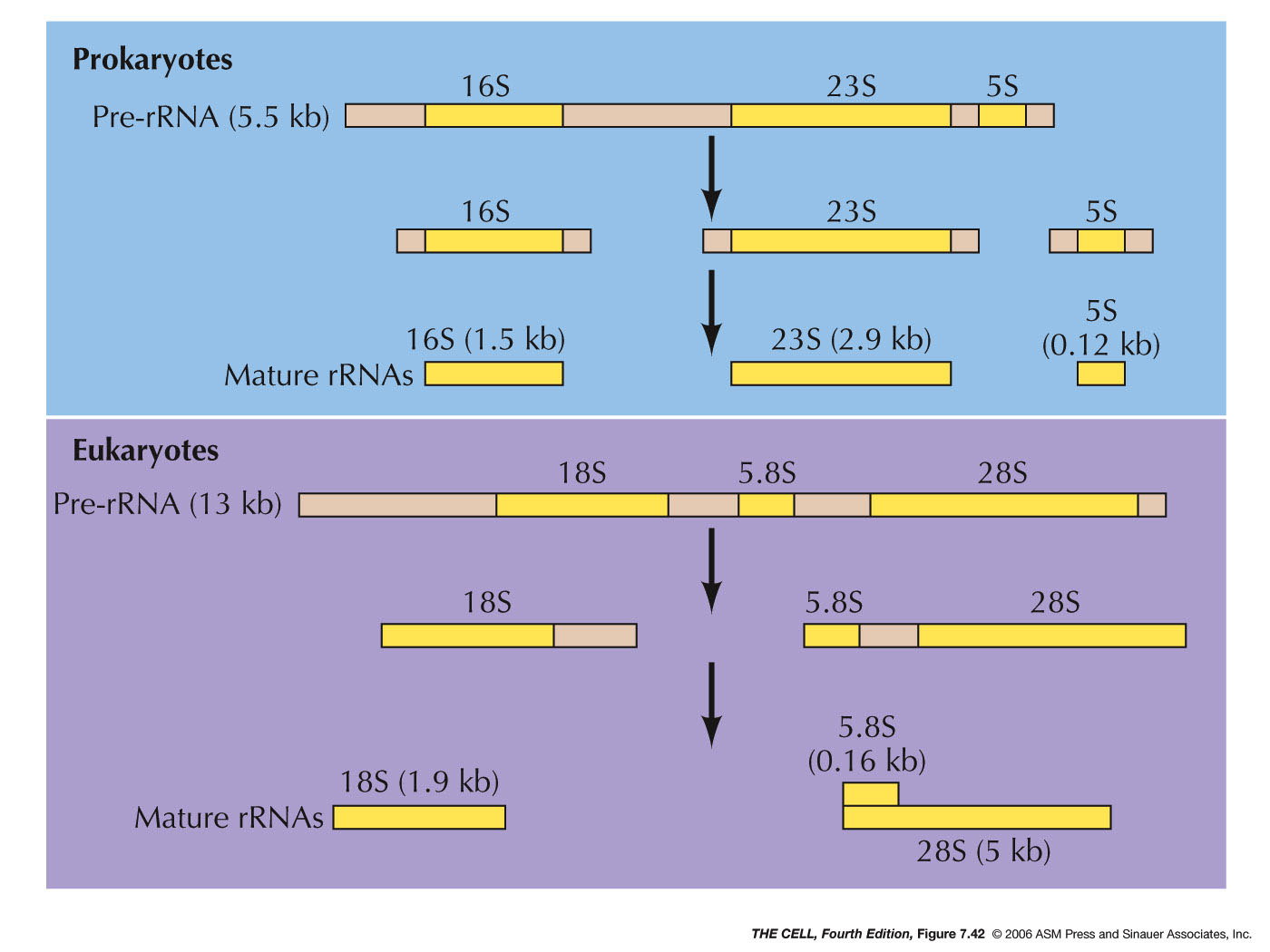
- rRNA Processing:
In eukaryotes, RNA polymerase I makes a large RNA
molecule called pre-rRNA (45S) which is subsequently
cut into three pieces yielding the 28S, 18S, and 5.8S
rRNA molecules. The small 5.8S molecule hydrogen bonds
to the end of the 28S molecule. (In E. coli, a
similar process produces three E. coli rRNAs.)(Ribosomal subunits) The
smallest eukaryote rRNA (5S) is made from a separate
gene by RNA polymerase III. All eukaryote rRNA genes
are tandemly repeated (up to several hundred times). (E. coli's rRNA
genes are also repeated, but only about 3-10 times.)
This repetition is presumably due to the fact that the
cell must be able to make a lot of this transcript
(rRNA) in order to make ribosomes. After rRNA
synthesis, the molecules are chemically modified by
methylation of some bases and of ribose. Also, uracil
may be modified into an altered base. These rRNA
modification occurs in the nucleolus (more later in
the course on this structure and the assembly of
ribosomes).
|
|
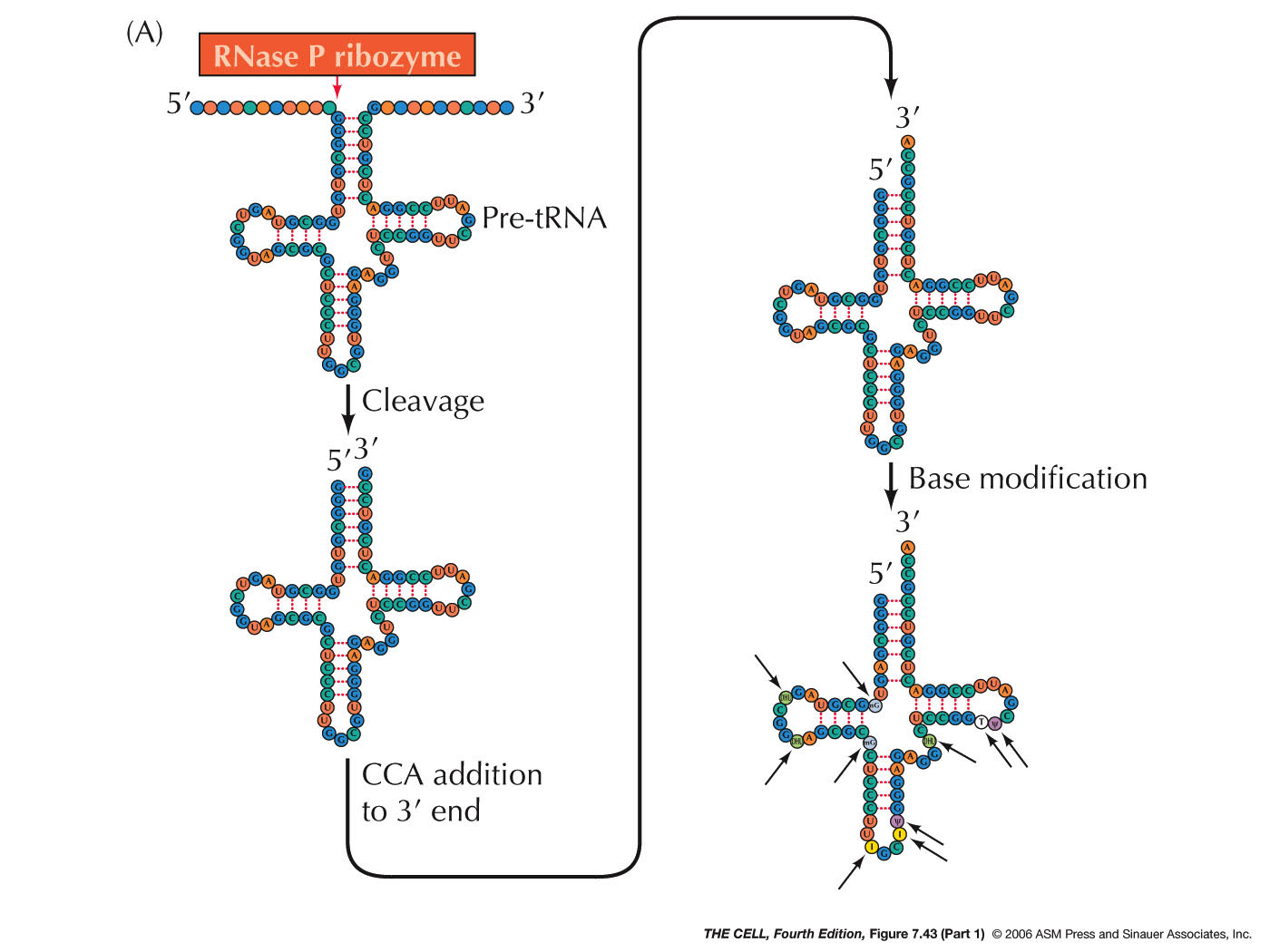
- tRNA Processing:
Eukaryotic tRNAs are made by RNA polymerase III. They
are usually made as long precursor molecules,
sometimes consisting of more than one tRNA. RNase P
cuts the pre-tRNA near the 5' end and another enzyme
cuts it near the 3' end (see 2010
article). RNase P is a complex of RNA and
protein and the surprising discovery of Altman in 1983
was that it was the RNA that was acting as the
catalyst (catalytic RNA, ribozyme). After the 3' end
is cut, another enzyme adds a CCA trinucleotide to the
3' end (if the CCA is not already there). tRNAs
undergo extensive base
modification which will be important in
translation. (Some tRNA have introns that must be
removed, but this is done by "normal" protein enzymes,
not catalytic RNAs -- see discussion on splicing.)
|
|
|
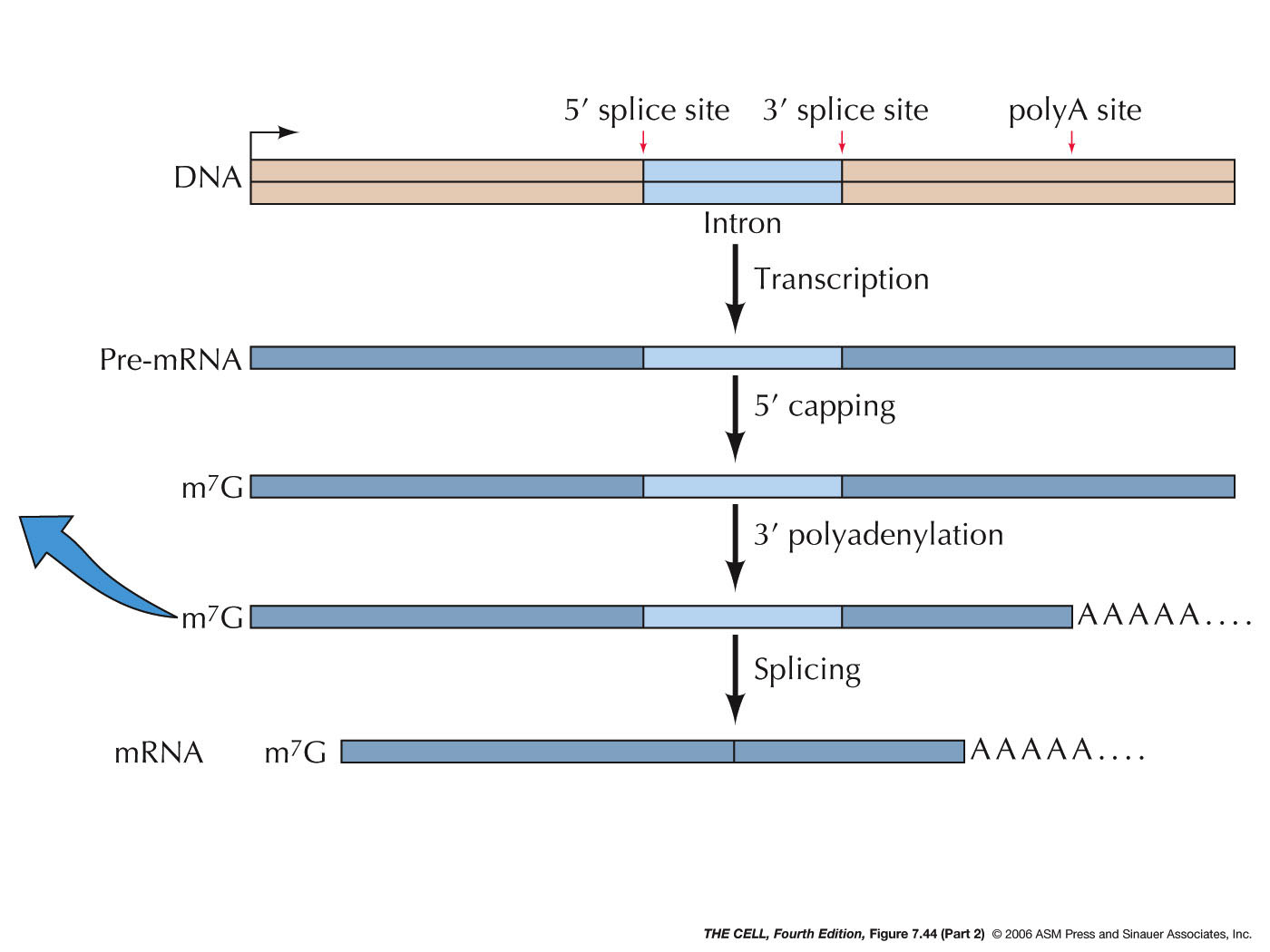
- mRNA Processing:
Unlike prokaryotic mRNAs, eukaryotic RNAs that code
for protein are not ready to begin the translation
process as soon as transcription ends (or in the
middle of it, as in prokaryotes). That is, they are
not yet "real" mRNAs. These pre-mRNAs (sometimes
called primary transcripts) are made by RNA polymerase
II. This RNA polymerase has a unique domain called the
C-terminus domain (CTD) where factors that process the
pre-mRNA bind. The other eukaryotic RNA polymerases (I
and III) do not have this CTD so they will not be
process the same way pre-mRNA are. Pre-mRNA processing
involves several events.
|
 |
|
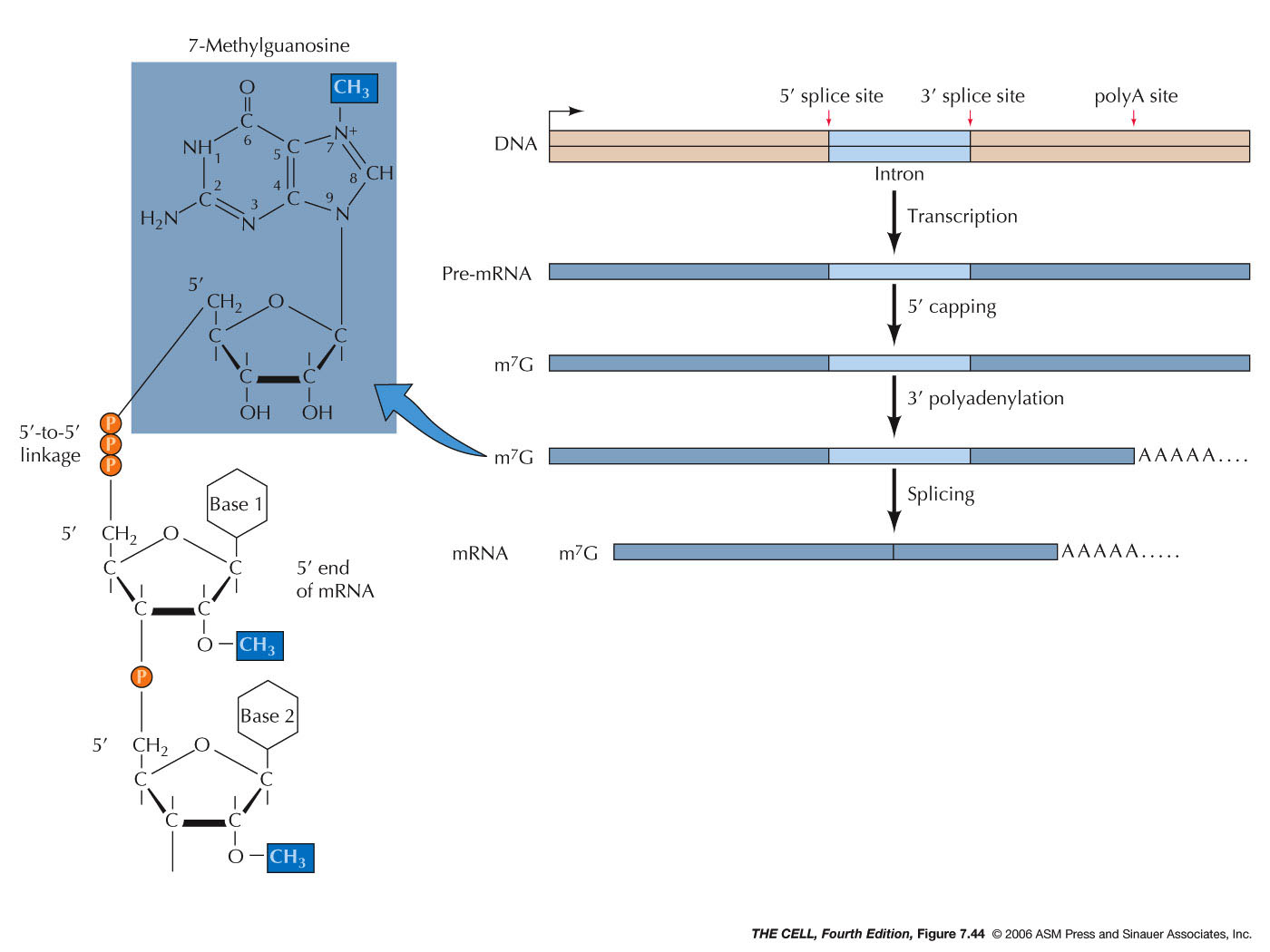
- 7-Methylguanosine
Capping: After about 20-30
nucleotides are synthesized, a GTP binds in reverse
configuration to the 5' end of the pre-mRNA. This is
then methylated. This methylguanosine cap stabilizes
(it cannot be digested by exonucleases) the RNA and
will be important in the translation process.
|
 |
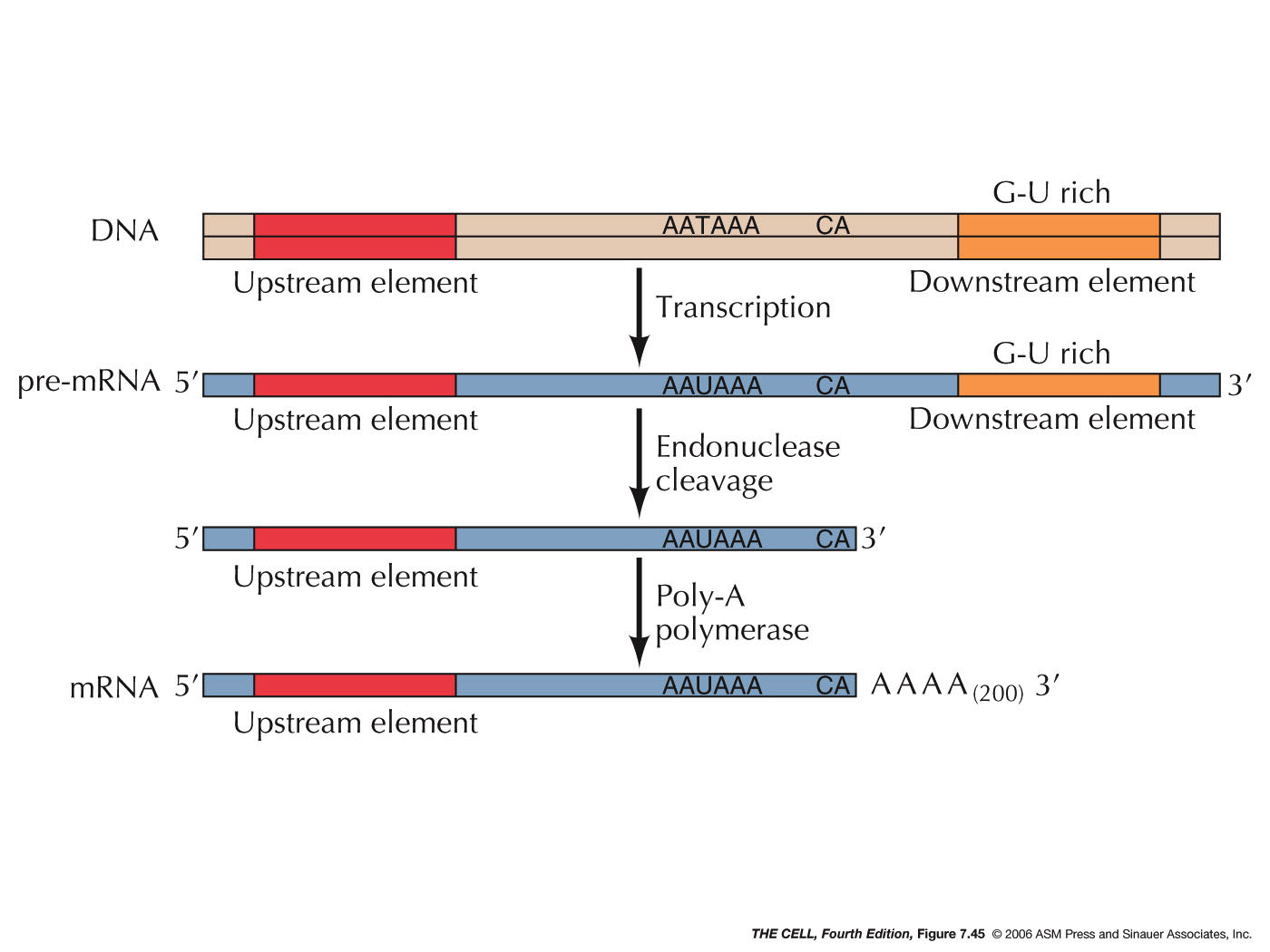
- Polyadenylation:
Another event is the enzymatic addition of
about 200 adenine nucleotides to the 3' end of the
pre-mRNA. The sequence AAUAAA occur about 10
nucleotides before a spot where there is a CA. A set
of protein factors associated with RNA polymerase
II's CTD cuts off the RNA on the 3' side of the CA
then this same set of proteins adds the poly-A tail.
The actual addition of the poly-A tail is by PAP
(polyadenylate polymerase), which is one of these
CTD-associated proteins. It uses ATP as the building
blocks and build the poly-A tail one nucleotide at
at time. This poly-A tail is needed for the mRNA's
transport to the cytoplasm (as a specific protein
binds to it causing its transport), for its
stability, and for the translation most mRNAs. The
polyadenylation/cleavage stimulates RNA polymerase
II to slip off of the DNA template thus terminating
transcription.
|
 |
- Splicing:
A 1977 discovery rocked the scientific world. Up
until that time, it was assumed that transcription
and translation in prokaryotes and eukaryotes was
essentially the same process. However, it was
discovered that the pre-mRNA molecule of eukaryotes
has several internal segment cut out and discarded.
The first clues came from the fact that mRNA is much
shorter than pre-mRNA. Furthermore, capping and
polyadenylation occur before the RNA is shortened
and these modifications remain in the finished mRNA.
So, how does it get shorter if the ends are
preserved? The answer came with the discovery of
introns (mRNA-DNA hybridization experiments).
Eukaryotic genes are transcribed into pre-mRNA and
then certain segments (introns) are snipped out and
discarded and the remaining segments (exons) are
spliced together. The average human gene has about 8
introns which make up over 80% of the gene (average
gene = 30 kb, average combined exon length = 2.5
kb).
|
- The Process of
Splicing: Splicing
occurs in two steps as listed below. (When
capping, polyadenylation, and splicing are
finished, we can legitimately call this molecule
mRNA. It will then be transported to the cytoplasm
for translation, exiting through the nuclear
pores.)
|
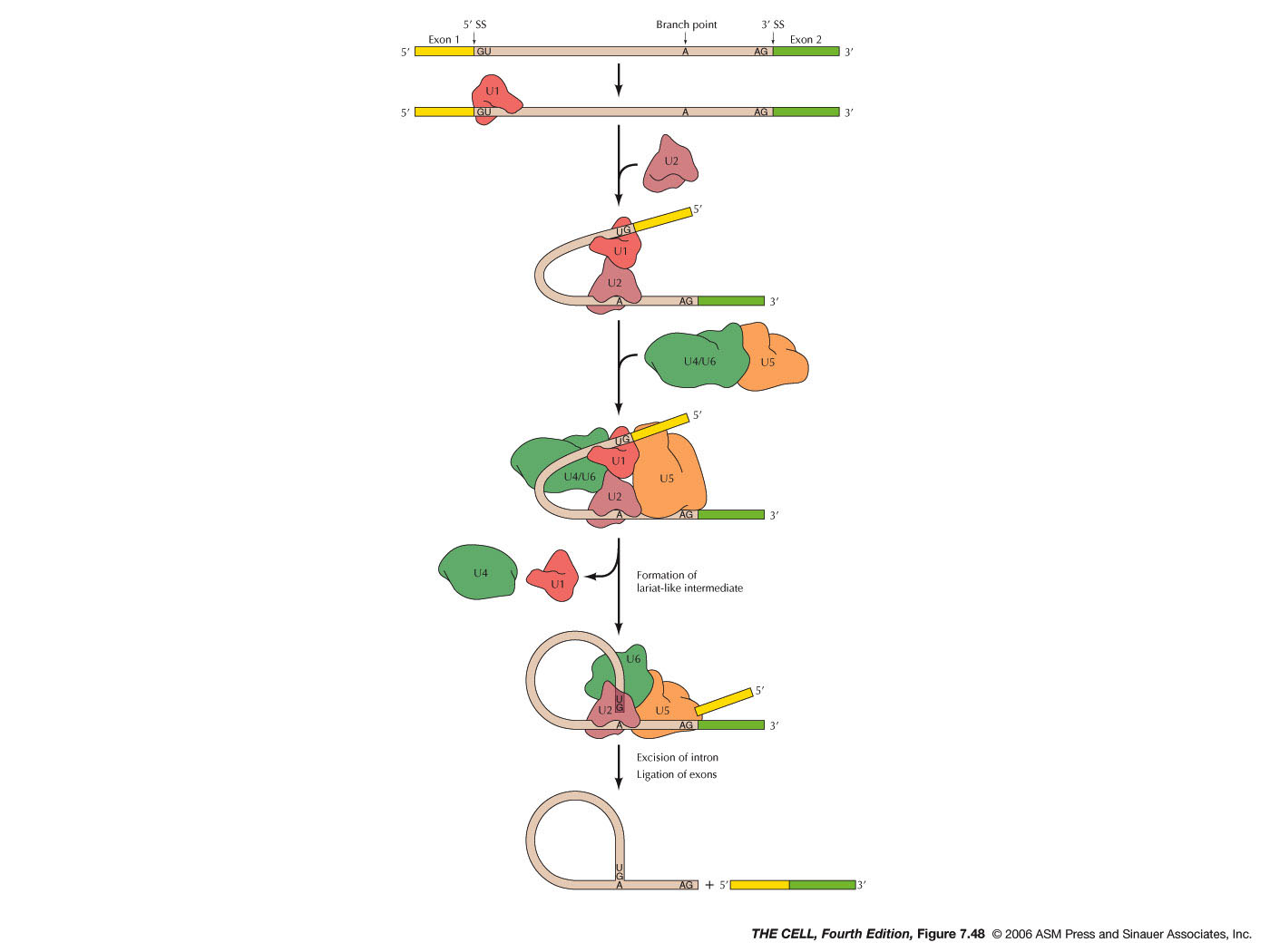 |
- Lariat
Formation: The pre-mRNA is cut at the
5' splice site (5' end of the intron) and
simultaneously that newly-cut 5' end binds to
the branch point adenine nucleotide (5' end
to 2'-OH of the A nucleotide). This forms a
lariat- (lasso-) shaped intermediate.
|
- Splicing:
The 3' splice site (3' end of the intron) is cut
and simultaneously the two exons are joined. The
lariat is then opened and degraded. These two
steps are catalyzed by the spliceosome which
includes proteins (in human there are probably
over 170 proteins) and small nuclear RNAs
(snRNAs, from 50 - 200 nt long, called U1, U2,
U4, U5, U6). A spliceosome therefore is a small
nuclear ribonucleoprotein (snRNP). Since U2
and U6 can catalyze splicing by
themselves, the catalytic activity of the
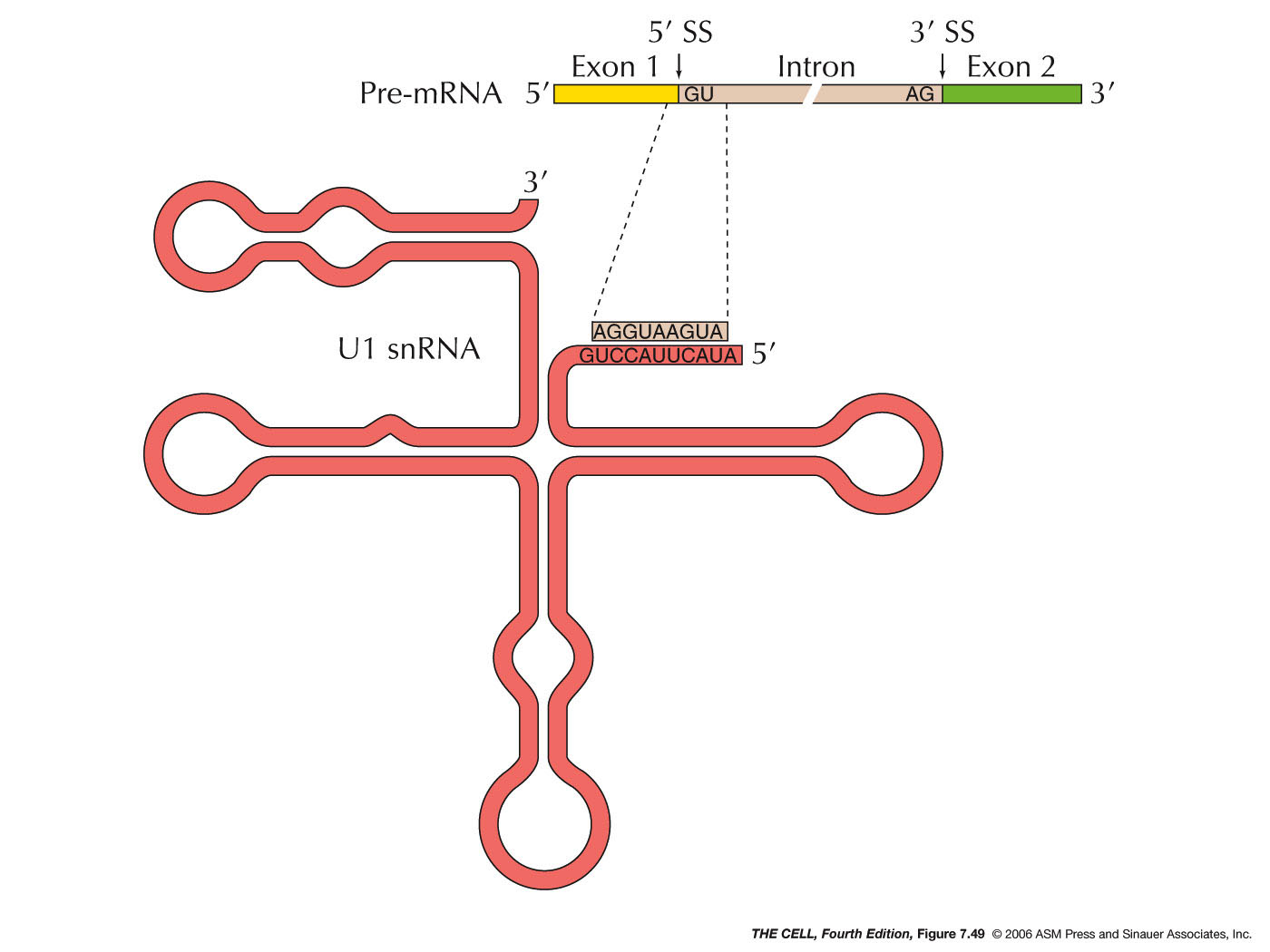
spliceosome is in the RNA (as with RNase P). A
short sequence at the 5' end of the U1 snRNA is
complementary to the 5' splice sites of the
pre-mRNA. During splicing, U1 binds first to
start the process. Another snRNA, U2,
and proteins bind with U2 base pairing at the
branch point. U6, U5, U4 and more
proteins bind. U6 appears to associate
with the 3' splice site and U2
associates with U6. There is evidence
that U5 associates with exon sequences
near both the 5' and 3' splice sites, possibly
tethering the exons together until they are
joined. (See this
page for an overview of the roles of the
snRNAs.)(Some self-splicing RNAs have been
discovered in Tetrahymena.)
- (Splicing
is no longer a eukaryote-only process!)
|
 |
- Transcription summary video
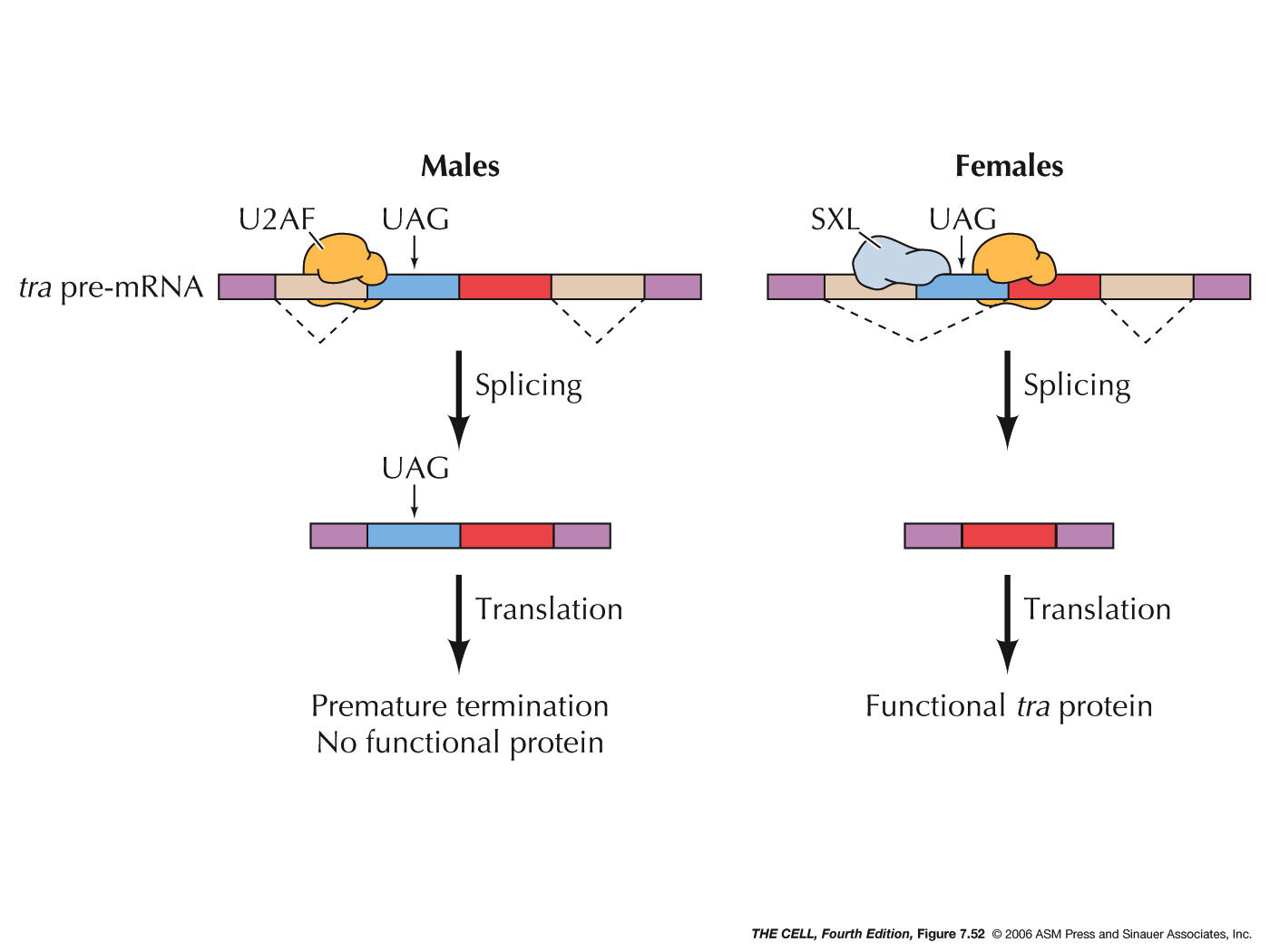
- Alternative
(Alternate) Splicing: Different introns
may be used at different times or in different
tissues. It is estimated that 50% of human genes
can be alternately spliced. One example occurs in
Drosophila
in which one
important sex-determining gene is
alternately spliced in males versus females. The
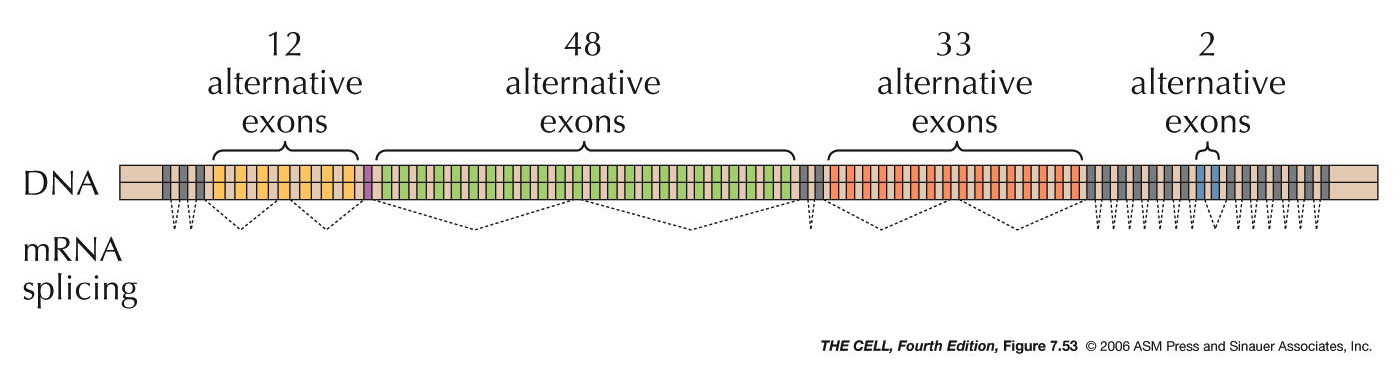
most incredible case found so far is also in Drosophila in
which the possible splicing
alternatives is unbelievably
high. (Meaning of splicing notation)
|
 |
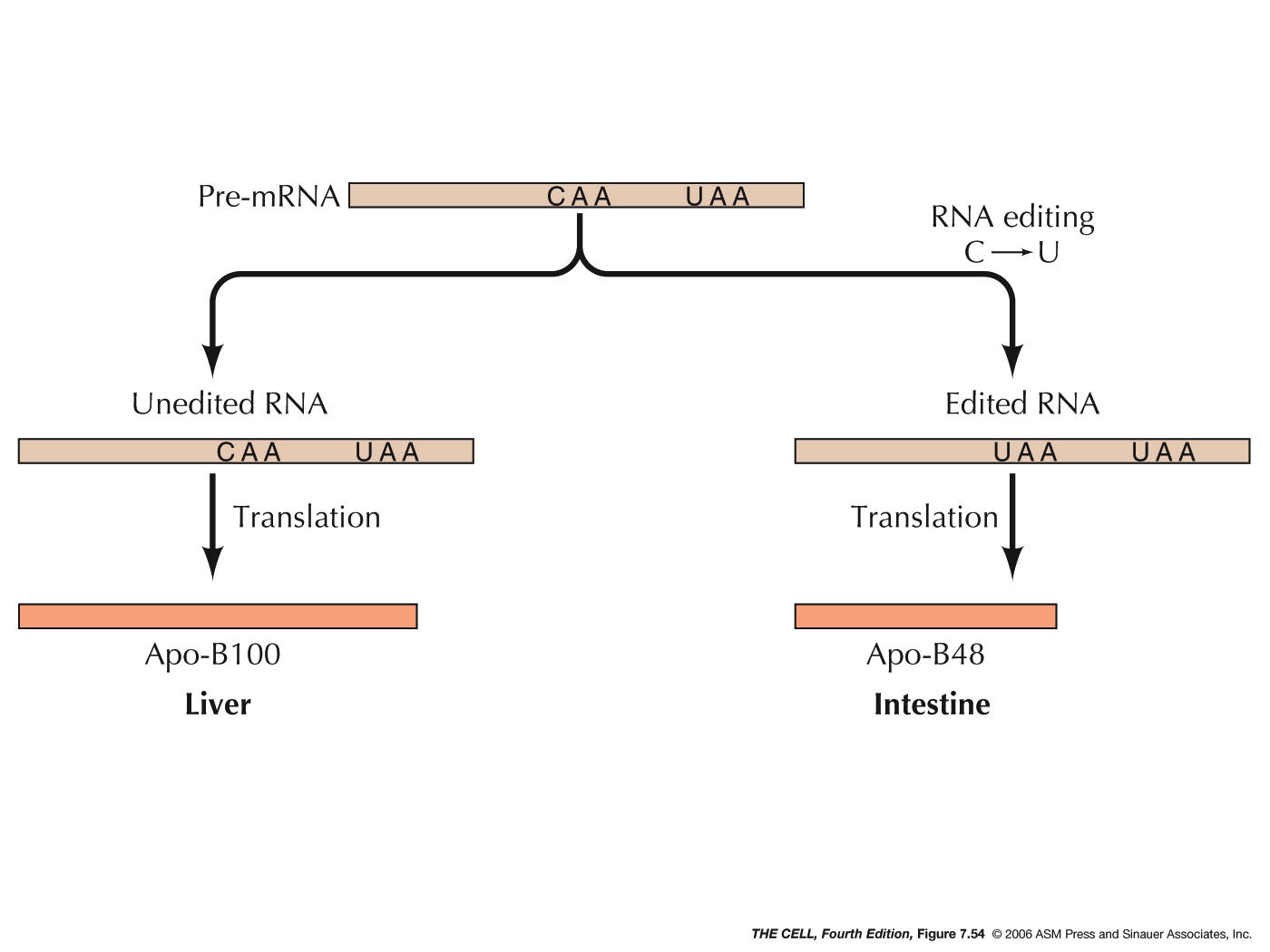
- RNA
Editing: RNA bases may be changed after
transcription, resulting in a change in the gene
product (amino acid sequence). In one case in
humans, apolipoprotein
B mRNA can have a base changed by editing that
results in the termination of translation (CAA
---> UAA, a nonsense codon). As a result, the
4536 amino acid-long protein found in liver is
shortened to a 2152 amino acid-long protein in the
intestine. Also, A ---> I changes are a common
mode of RNA editing. (Serotonin receptor mRNA can be
edited at 5 sites.)
|
 |
- Squid are
hyper-editors when it comes to RNA (by
Lisa D. Chong, Science Feb 27, 2015):
During RNA editing, specific enzymes alter
nucleotides in mRNA transcripts so that the
resulting protein differs in amino acid sequence
from what was encoded by the original DNA. Such
RNA editing is a means to generate greater protein
diversity; however, most organisms only use it
sparingly. Alon et al. (eLife 4,
e05198, 2015), however, now report an exception.
They sequenced RNA and DNA from the squid nervous
system and discovered that 60% of the transcripts
exhibited RNA editing. Such "recoding" occurred
largely in genes with cytoskeletal or neuronal
functions and may be advantageous to organisms
such as squid that must respond quickly and
continually to environmental changes.
|
- RNA
Degradation: The expression of a gene is a
function of the rate of production of its mRNA and
the rate of degradation of that mRNA (among other
things). Therefore, RNA degradation is an important
process. Cells have a nonsense-mediated mRNA decay
process which destroys mRNA that have a premature
nonsense codon. "Old" mRNA are routinely degraded.
The half life of prokaryotic mRNA is about 2-3
minutes versus 30 minutes to about 20 hours in
eukaryotes. (mRNA
degradation)(measuring
RNA synthesis and degradation)
|
 Home
Home




















