Transcriptional Controls: The
main mechanism of turning genes on and off occurs at the
level of transcription. We have already seen at least one
site that although it is outside the coding sequence of the
gene, is important in proper proper gene expression: the
promoter (RNA holoenzyme binding site). However, other sites
are also involved.
- Transcriptional
Regulation
in
E. coli: One Example: The regulation of the
genes involved with lactose catabolism in E. coli was
discovered by Jacob and Monod in 1961. E. coli only
produces (in any significant quantity) a set of enzymes
necessary to break down lactose when lactose is present.
- The
lac
Operon: The genes that encode the three
protein needed for lactose catabolism lie adjacent to
each other. These three genes plus the regulatory
sequences nearby constitute the lac operon. The three
genes are transcribed as one polycistronic message,
therefore if one is turned on, all three are turned
on. (This kind of clustering of functionally related
genes is seen often in prokaryotes.)
|
- The Structural Genes: The enzymes
are β-galactosidase
(cleaves lactose), lactose permease (facilitates the
entry of lactose into the cell), and transacetylase
(gets rid of a tag-along toxin that enters with
lactose by the action of lactose permease). The
genes that code for these three proteins are
designated z,
y, and a,
respectively.
|
 |
- The Promoter: This is the RNA
polymerase holoenzyme binding site (as described
earlier).
- The Operator: This site is a
sequence to which the repressor protein binds. It is
just downstream from the promoter. When repressor
protein is bound, RNA polymerase is prevented from
binding to the promoter. This sequence is designed o. The operator
was discover by finding cis-acting constitutive mutants
(always produced the three gene produces, regardless
of the presence or absence of lactose or repressor
protein).
- The Repressor: This is a protein
coding sequence which produces the repressor
protein, capable of binding to the operator (and
turning off transcription). This gene is designated
i. It was
discovered as a trans-acting
gene that apparently made a diffusible substance
which could affect an operator on a different
"chromosome." (merodiploid, or merozygote F' cells)
- CAP Binding Site: This site is about
60 nucleotides upstream from the transcription start
site. It binds a protein called catabolite activator
protein (CAP). The binding of cAMP to CAP stimulates
it to bind to the CAP binding site. This binding
then facilitates the binding of RNA polymerase to
the promoter (enabling transcription).
|
- Negative Control in the lac
Operon: When lactose is not present,
the level of transcription of these three genes is
negligible. However, in the presence of lactose, their
transcription is greatly ramped up. This process is an
example of negative control, since there is something
that normally turns the expression of this gene off
(which can be regulated). In the "normal" state (no
lactose present), the i gene makes repressor protein which
binds to the o
site, preventing RNA polymerase binding, thereby
turning off transcription. However, in the presence of
lactose, an isomer of lactose (allolactose) binds
allosterically to the repressor, preventing it from
binding to o.
Therefore, RNA polymerase can bind to the promoter and
transcription of these three genes is turned on.
(Protein binding turns off the operon = negative
control) lac
operon problems:
Lac Operon Problems:
- I+ is a normal
repressor gene
- I- is a mutant repressor gene
that does not make any functional repressor
protein.
- IS is
a mutant of the Lac repressor gene which makes
repressor protein that represses the Lac
operon even in the presence of inducer
(lactose)(it makes a repressor protein to which
allolactose cannot bind).
- O+ is a normal
operator sequence
- O- is an mutant
operator to which repressor protein cannot bind.
- P+ is a normal
promoter sequence
- P- is a promoter
sequence to which RNA polymerase cannot bind.
- Z+ makes normal,
functional β-galactosidase
- Z- is a mutant that
makes no functional β-galactosidase
- Y+ makes normal,
functional lactose permease
- Y- is a mutant that
makes no functional lactose
permease
For each of these genotypes, indicate
whether β-galactosidase and lactose permease would be
produced constitutively, inducibly, or not at all.
(What does constitutively and inducibly mean?)
I- P- O- Z+
Y+
/ I+
P+
O+
Z+
Y-
I-
P+
O-
Z+
Y+
/ I+
P+
O+
Z-
Y-
I- P+ O- Z+
Y+
/ I+
P+
O+
Z-
Y-
I- P+ O- Z+
Y+
/ I+
P+
O+
Z+
Y+
I+
P+
O+
Z+
Y+
/ I+
P+
O+
Z-
Y+
I-
P+
O+
Z-
Y+
/ I+
P+
O+
Z+
Y+
I+
P+
O+
Z+
Y-
/ I- P+ O+ Z+ Y+
IS P+
O+
Z+
Y+
/ I+
P+
O+
Z+
Y+
I+
P+
O-
Z+
Y-
/ I+
P+
O+
Z-
Y+
I+
P-
O+
Z+
Y+
/ I+
P+
O+
Z+
Y+
|
 |
- Positive Control in the lac
Operon: One wrinkle in this situation
is that if both glucose and lactose are present, the
cell apparently "prefers" to use glucose over lactose,
and the lac operon is turned off. This is positive
control because the event involved is the facilitation
of transcription. When the concentration of glucose is
high, the concentration of cAMP is low (due to the
regulation of another gene: adenylyl cyclase,
which converts ATP to cAMP). When glucose levels drop,
cAMP levels rise. Therefore, when there is a high
concentration of glucose, there is little cAMP, so it
cannot bind to CAP. Without cAMP, CAP does not bind to
the CAP binding site, and therefore, RNA polymerase
binding is not facilitated. Transcription is off. But,
if there is little glucose, then cAMP levels are high,
so CAP-cAMP levels are high, and CAP-cAMP's
binding to the CAP binding site facilitates RNA
polymerase binding to the promoter, turning
transcription on. (Binding turns on.) So, for the lac
operon to be turned on, lactose must be present and
glucose absent (or in low concentration). That is, the
negative control mechanism has be inactivated and the
positive control mechanisms has to be activated. Many
similarly regulated mechanisms have been found in
prokaryotes.
- (About how CAP-cAMP facilitates RNA polymerase
binding to the promoter: CAP "recruits" RNA polymerase
to bind to the promoter. It interacts with a few amino
acids on the RNA polymerase α-chain CTD and causes
them to bind to the promoter, thus causing the whole
RNA polymerase to bind to the promoter. The reference
is here.
Start reading at "Transcription activation at class I
CAP-dependent promoters.")
|
 |
- Transcriptional
Control
in
Eukaryotes: While there are many common
features between prokaryotic and eukaryotic gene
regulation (like the presence of a promoter and the
presence of controlling sequences upstream from the
structural gene), there are some major differences. One
is that eukaryotes do not have polycistronic message and
we usually do not see clusters of functionally related
genes that are turned on by turning on the transcription
of a single RNA. Functionally related genes are often
scattered across the genome and therefore co-regulation
is more complex than in prokaryotes.
|
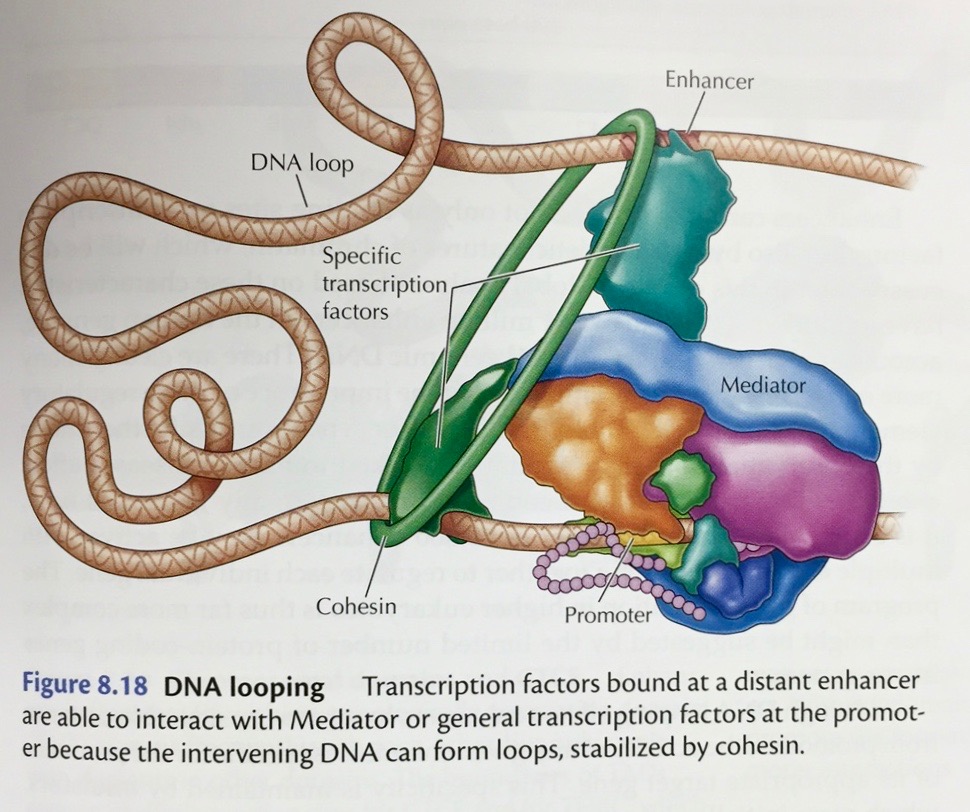
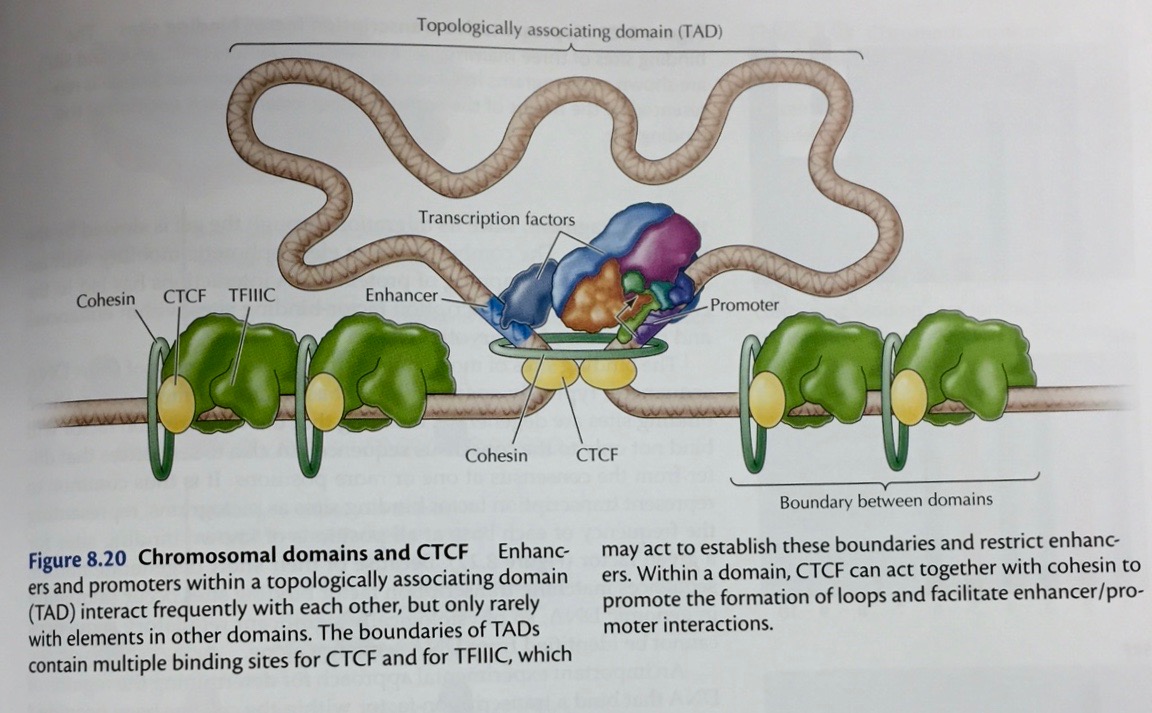
|
- Repressors:
Proteins may also bind to sites near the promoter and
interfere with RNA polymerase binding. They may also
interact directly with RNA polymerase to prevent
transcription.
|
 |
- Chromatin
Structure
and
Transcription: Transcriptionally
active
chromatin is not highly condense (it is not
heterochromatin), but apparently in the 30 nm fiber
condition. The nucleosomal histones must be removed
for transcription. This may be by HMGN protein binding
(similar to H1) causing decondensation, or by
acetylation of histones.
|
 |
- Noncoding
RNAs: These small RNAs can regulate gene
expression post-transcriptionally, but they can also
inhibit transcription by causing methylation of
histones and thus the formation of heterochromatin.
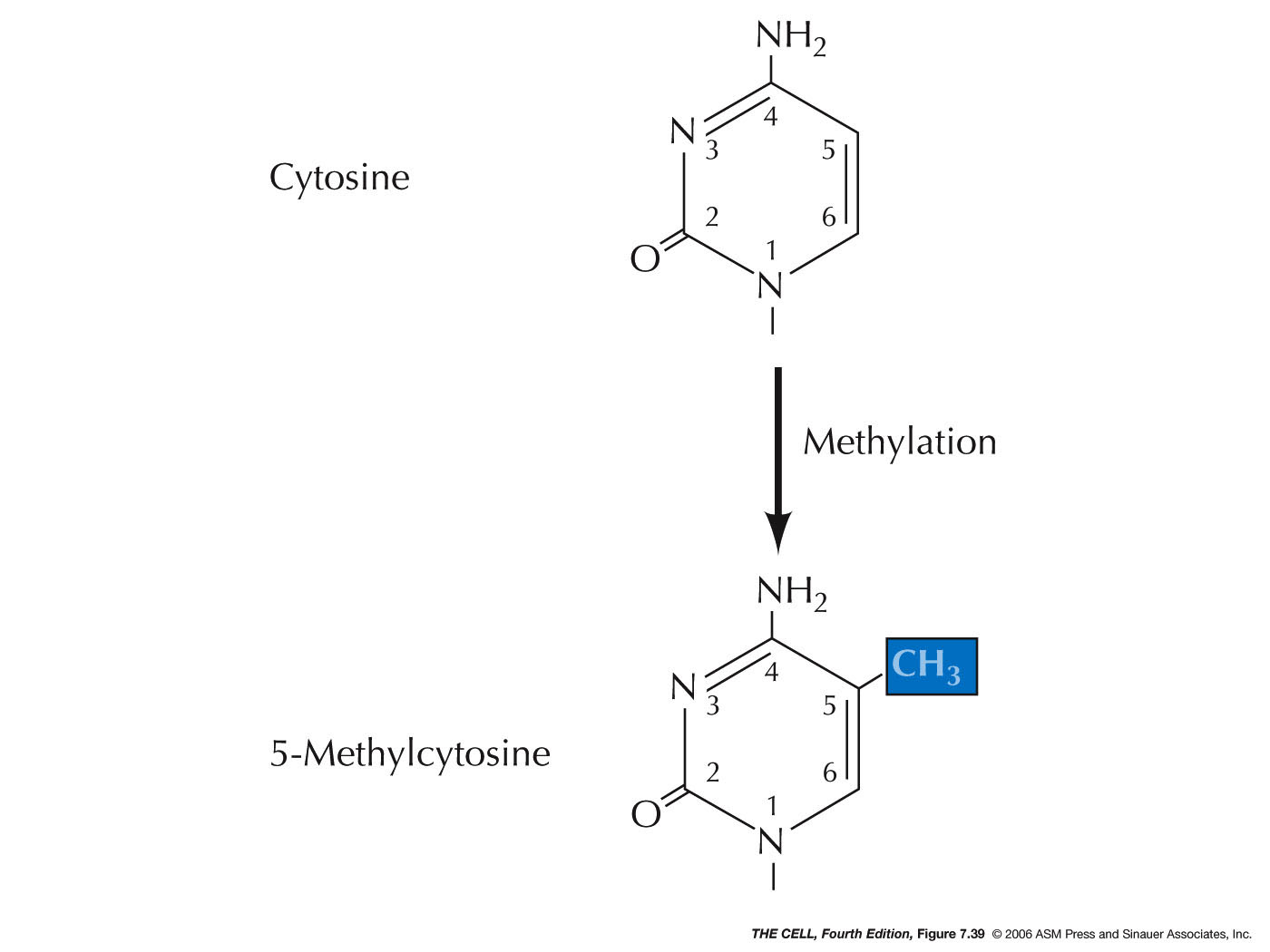
- Methylation
and Epigenetics: In
vertebrates, the addition of a methyl group to a
base (especially cytosine
forming 5-methylcytosine) is often
involved in transcriptional inactivation. Enzymes
that add methyl groups to DNA are called DNA
methyltransferases (DNMTs). Methylation patterns are
generally preserved after replication by DNMTs.
Methylation of DNA can turn off transcription by
altering histones (causing chromatin compacting) and
may also affect the binding of specific
transcription factors. Unmethylated CpG islands (see
"Part 3" video above) show hyperacetylation
of histones H3 and H4, a deficiency in histone H1,
with nucleosome-free regions promoters
(transcription is on). (Methylation
Video details will not be on the test--except
the summary covered in the "Part 3" video above.)
Epigenetics: 2010 article.
An interesting example is seen in the rare phenomenon
of gene imprinting (unusual
event of methylations being NOT removed during
zygote formation) as occur in the H19 gene.
(Imprinting/methylation is an example of epigenetic
changes to the gene: 2010 article)
This gene produces only an RNA product with function
yet unknown (but may related to cancer: 2010 article)(Methylation and
resetting the genome)
|
|
-
CpG density
defines two classes of RNA polymerase II
promoters
Despite the
sequence diversity among promoters, genes
transcribed by RNA polymerase II can be
classified in two different and mutually
exclusive groups according to the distribution
of CpG dinucleotides across their 5' ends. In
one class, the frequency of CpGs is the same as
the genome average, which is roughly one every
100 nucleotides. This class invariably includes
genes whose expression is restricted to a
limited number of cell types... In contrast, the
5' end of the genes belonging to the other group
is surrounded by a region ~1 kb long where the
frequency of CpGs is approximately 10 times
higher than the genome average. These regions
were very appropriately called CpG islands
... and show such a conspicuous clustering of
CpG dinucleotides that it can be readily
detected by visual inspection of the CpG plot.
The consistent association of CpG islands with
the upstream region of many genes immediately
suggested a possible involvement in
transcriptional regulation ....
What is so special about CpGs relative to the
other 15 possible dinucleotides in DNA? CpGs are
the sites where methylation takes place, and ~80
% of them are methylated at position 5 of the
cytosine ring in humans and mice. Somewhat
paradoxically, CpGs
remain nonmethylated at CpG islands,
despite their abundance, whereas the majority of
the remaining CpGs scattered across the genome
are mostly methylated. In addition to the lack
of methylation, human and mouse CpG islands have
a G+C content of 67 and 64 %, approximately,
while the genome averages are 41 % and 42 %,
respectively...
These distinctive features of the CpG islands in
terms of a lack of methylation and an elevated
G+C content are accompanied by an equally
distinctive chromatin organization. Chromatin
analysis at global genomic level has revealed
that CpG islands show the properties usually
ascribed to "open" or "active" chromatin.
This includes hyperacetylation of histones H3
and H4, a deficiency in histone H1, positioned
nucleosomes and nucleosome-free regions that
coincide with enhanced sensitivity to nucleases
relative to bulk DNA... These properties
highlight CpG islands as regions that are
particularly well suited for direct access to
DNA, which is consistent with their
co-localization with the promoters of
many genes. Previous studies based on the
biochemical isolation of the CpG island fraction
estimated approximately 45,000 and 37,000 in the
human and mouse genomes, respectively...,
although recent computational predictions have
lowered these figures to about 27,000 and
15,500...
Regardless of the absolute number of CpG islands
in the genome, a more relevant issue is what
kind of genes are associated with them, since
only ~60% of all human genes are associated with
CpG isands. This includes all the
housekeeping genes--those expressed in
all cell types--and about half of the tissue-
specific genes...
Transcription
from CpG-rich and CpG-poor promoters
How does methylation affect transcription from
CpG island and non-island promoters? Since CpG
islands are nonmethylated in sperm and remain
consistently devoid of methylation in somatic
tissues, regardless of the expression of the
genes associated with them, it is unlikely that
DNA methylation would play any role in their
regulation. Exceptions to this rule are the CpG
islands of imprinted
genes, those in the mammalian X
inactive chromosome and those associated with
the MAGE genes that become methylated during
normal mammalian development... Even in this
case, methylation is not the primary
inactivating signal but takes place at a stage
when transcription has been switched off by
other means...
Transcription is strongly repressed upon
unscheduled de
novo methylation of CpG islands in cell
lines and tumour cells, a phenomenon that occurs
at high frequency in these situations but never
in the organism under normal physiological
conditions with the exceptions mentioned
above...
In contrast with CpG islands, CpG-poor promoters
are methylated in sperm and are always
associated with tissue-specific genes.
A direct role of DNA methylation in the
regulation of this class of promoters predicts a
correlation between their methylation profile
and their level of expression. Many examples and
also several exceptions to this correlation have
been described, suggesting that although DNA
methylation affects gene expression, it is
unlikely to play a general role as a
transcriptional regulator... Detailed analysis
of the kinetics of demethylation and gene
expression has shown that in some cases where
demethylation correlates with expression, the
former fol-lows the binding of transcription
factors rather than being a prerequisite for
it. For example, binding of nuclear
factor kappa B (NF-kB) transcription factor to
an intronic enhancer of a k-chain gene is
required for demethylation in B cells...
That DNA
methylation, despite its prominent presence in
the mammalian genome, is not a general
regulator of gene expression is not surprising
since proper gene regulation takes place in
invertebrates, many of whose genomes have a
low or undetectable level of methylation.
|
 Home
Home







