 Home
Home
![]()
Department
of Biological and Environmental Sciences
Cell & Molecular Biology
Dr. David A. Johnson
Biol 405 4 Credits Spring
2017 MWF 11:45-12:50 AM PH 204
A Brief Overview of Cells and Cell
Biology
(ch.1 pp. 1-12, 17-38)
|
Living things are
composed of cells. Cells of archaea (domain: archaea) and
bacteria (domain: bacteria) are quite different from those
of the rest of the living world. They make up the
prokaryotes, while the rest (from protozoa to you) are
eukaryotes (domain: eukarya).
E. coli
Prokaryotes: Prokaryotes include the archaea
(many inhabiting harsh environments) and bacteria (including the
photosynthetic cyanobacteria). They lack a nucleus (no
nuclear membrane) but have a region called a nucleoid.
They have ribosomes but lack other cellular organelles.
The prokaryote chromosome is a "naked" circular DNA
molecule. As we will see in detail later, they also lack
the process of splicing. Eukaryotes:
Eukaryotes have a double nuclear membrane separating the
chromosomes from the rest of the cell. (Plant cell, animal
cell) Their chromosomes are linear DNA molecules
bound to protein (the most abundant protein being
histones). The process of splicing is seen only in
eukaryotes. In addition to ribosomes, they have a variety
of organelles and structures including these and others
(details later):
Origin of Cells: The
first cell are thought to have arisen from the pre-biotic
mixture in the oceans. Miller has
proposed how this early chemical evolution may have
occurred. Phospholipid molecules will self-assemble into a
bilayer. RNA has recently been
show to be able to catalyze its own replication, leading
to the hypothesis of an early, RNA world (with the more
stable DNA later replacing RNA as the genetic material of
most organisms)(an
alternative view to the RNA world - also pasted
below)(Still another
alternative view of the origin of life on earth!!!)(Newer
evidence for the RNA world.) Origin of Eukaryotes: Eukaryotes apparently arose after endosymbiosis involving a bacterium capable of oxidative phosphorylation (becoming mitochondria) and endosymbiosis of a cyanobacterium capable of photosynthesis (becoming chloroplasts). Evidence for this theory includes the structure of the DNA and the ribosomes of mitochondria. Experimental Cell Models:
Several model system have proven useful in understanding
cellular processes including these:


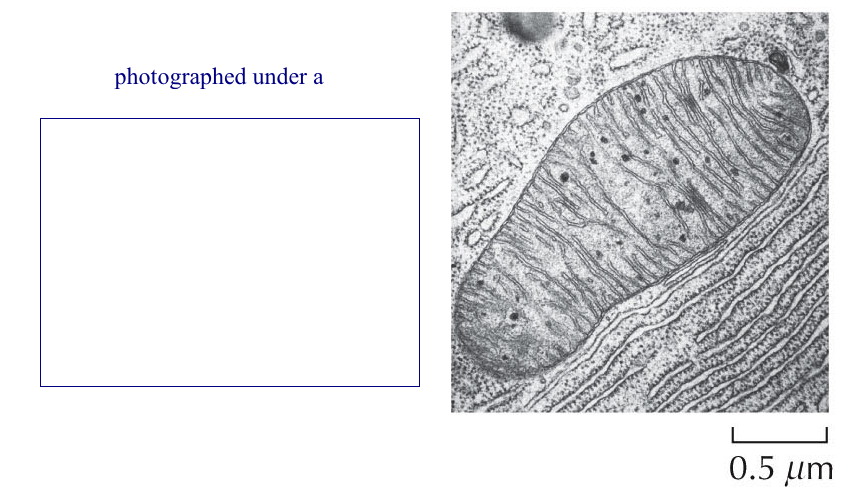
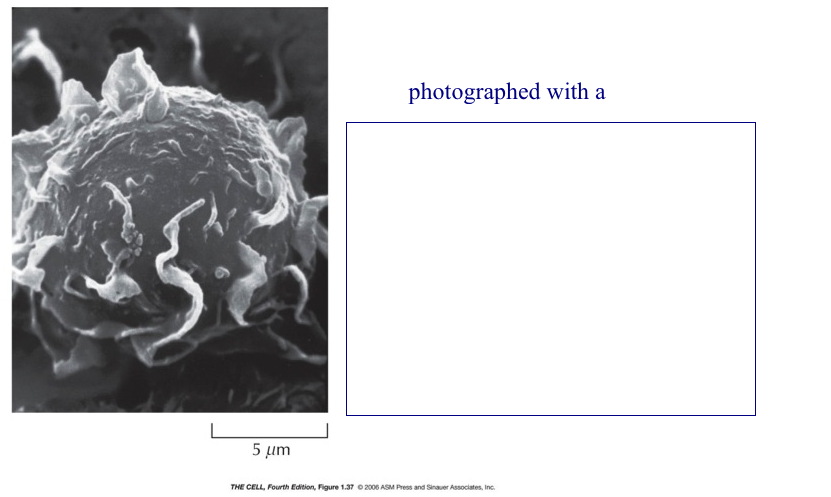
Microscopy: There are
at least two major types of microscopes.
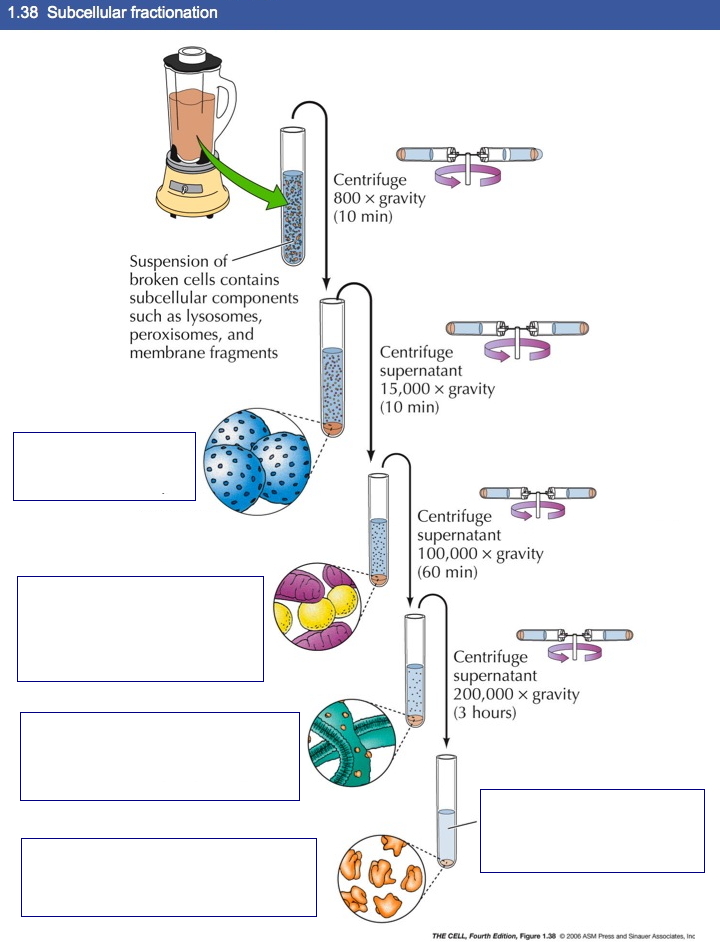
  Fractionation and Separation
of Cell Components: Cell components can be
separated by several methods.
Cell Culture: Primary and secondary cultures of
human cells will eventually stop dividing. Stem cell line
cultures (as well as cancer cell lines) keep dividing.
(January, 2012 news: Using
embryonic stem cells to treat macular degeneration?)(The
book "The Immortal Life of Henrietta
Lacks" Published by Random House is available in
paperback.)
|
|
(The material below will not be on any
exam.) (Stolen
from Dr. Daniel P. Heruth, former chair, Biology
Department, William Jewell College, Liberty, MO)
The
process of life is played out within the cell.
Therefore an understanding of the dimensions of the
cell and its contents is important to the study of
cell and molecular biology. It is important to realize
that the cell is a very crowded place, and thus very
organized. A typical eukaryotic cell has a diameter of
25 μm. If we magnified the cell by a million fold, we
would have a cell 25M in diameter. Twenty-five meters
is approximately 81 feet , so if we were standing in
the center of the cell it would be 40 feet in every
direction. Our magnified cell would be approximately
the size of a large college lecture room, if we
removed the seating. A water molecule, the most common
molecule within a cell, has a diameter of 0.4 nm.
Magnified a million-fold, the water molecule would be
only 0.4 mm in diameter. This is the size of the
period at the end of this sentence. Amino acids, the
building blocks of proteins are larger than water
molecules. They have an average molecular weight of
110 and are from 0.6 nm - 1.2 nm in length. Increased
by a factor of a million, the amino acids would be 0.6
mm - 1.2 mm in length or about three times larger than
the period. Even at this scale, we would not be able
to distinguish the atoms within the amino acid.
Hemoglobin, a protein made from amino acids has a
molecular weight of 64,000 and a diameter of 6.4 nm.
Magnified it would have a diameter of 6.4 mm or the
size of a pea. Ribosomes, large complexes of proteins
and RNA, has a diameter of 25 nm. Magnified a
million-fold would increase the diameter to 25 mm, the
size of a ping pong ball. In a typical eukaryotic cell
there are more than 20,000 ribosomes. A mitochondrial
organelle, the site of ATP synthesis, is 1μm in
length; amplified it would become a 1 m long oversized
football, much like a rugby ball. A typical liver
cell, a hepatocyte contains 800-2500 mitochondria
(~20% cell volume). A chloroplast is 5X larger than a
mitochondrion, so if our cell was a plant cell you can
imagine that we may be running out of room. DNA is 2
nm wide, so in our cell it would be a 2 mm wide strand
of sewing thread. A human cell contains 1.8 m of DNA.
Therefore, if we were to stretch out all of the DNA in
our magnified cell end to end we could extend it over
19,500 football fields. Remarkably, this DNA would
need to be coiled around histone proteins and packaged
tightly within the nucleus, a membrane bound region
found in the center of our cell. This is possible
because DNA is a very thin molecule. Even more
remarkable is the plasma membrane which surrounds and
protects the cell. Magnified a million-fold, the
membrane would be only 1 cm thick. How can such a thin
membrane complex of proteins and lipids define the
fundamental unit of life? Welcome to the world of the cell! New
Study Contradicts RNA World Theory BUT: Some even newer evidence says maybe the RNA world idea is right after all: Shedding
Light on the Origins of Life According
to the RNA world hypothesis, self-replicating RNA
molecules were the precursors to all current life on
Earth. Ancestral organisms contained RNA genes that
were replicated by RNA enzymes, and it was only later
that genetic information and catalytic functions were
transferred to DNA and proteins, respectively.
Horning and Joyce moved closer to that goal in a
recent study published in Proceedings of the National
Academy of Sciences (PNAS) [1]. They reported the in
vitro evolution of an improved RNA polymerase ribozyme
that can synthesize structured functional RNAs and
replicate short RNA sequences in a protein-free form
of PCR. According to the authors, the new findings
demonstrate that the two prerequisites of Darwinian
life--replication of genetic information and its
conversion into functional molecules--can now be
accomplished with RNA in the complete absence of
proteins.
According to Horning, this research could have a
variety of practical applications. "First,
we are able to amplify RNA by PCR directly, rather
than converting it into DNA, which could be useful in
studying or modifying biological RNA," Horning
said. "The polymerase may also be
tweaked to accept non-natural nucleic acids, which
might allow us to more readily use a variety of more
arcane nucleic acid variants which have proven useful
in biotechnology." |