"...alanine tRNA appears to
recognize GCU, GCC and GCA ... I have suggested that
this is because of a "wobble" in the pairing in the
third place..."
Francis H. C.
Crick (1966)
|
| The genetic information in
the form of a sequence of bases in DNA is transcribed
into individual "messages" in the form of a sequence of
bases in RNA. This process of transcription is followed
by the process of translation, in which the meaning of
this informational RNA molecule is decoded. Translation
is then the process by which an RNA base sequence is
used to directed the synthesis of a specific polypeptide
with a specific amino acid sequence. In eukaryotes, it
follows RNA processing and occurs after the mRNA leaves
the nucleus through a nuclear pore and travels to the
cytoplasm. That is, the two processes, transcription and
translation, are temporally and spatially separated. In
prokaryotes, however, translation begins before
transcription is finished (they occur simultaneously)
and they occur in the same place. In order for
translation to occur, several things must be
present--the major ones being an mRNA molecule, a
ribosome, and charged transfer RNAs (tRNAs). |
- Transfer RNAs:
These single-stranded RNAs are 70-80 bases in length
and all have common (conserved) sequences. There are
numerous tRNAs--at least one for every amino acid.
Eukaryotic and prokaryotic tRNAs are very similar in
many respects. tRNAs have many modified bases, as we
learned in the last unit. An amino acid will become
covalently bonded to its specific tRNA creating a
charged tRNA. Each tRNA is designated with a
superscript that indicates which amino acid it will
bind. For example, tRNAala
is the tRNA that will be charged with (bound to)
alanine. When it is charged, it is indicated as
ala-tRNAala (alanine
attached to the alanine-specific tRNA).
|
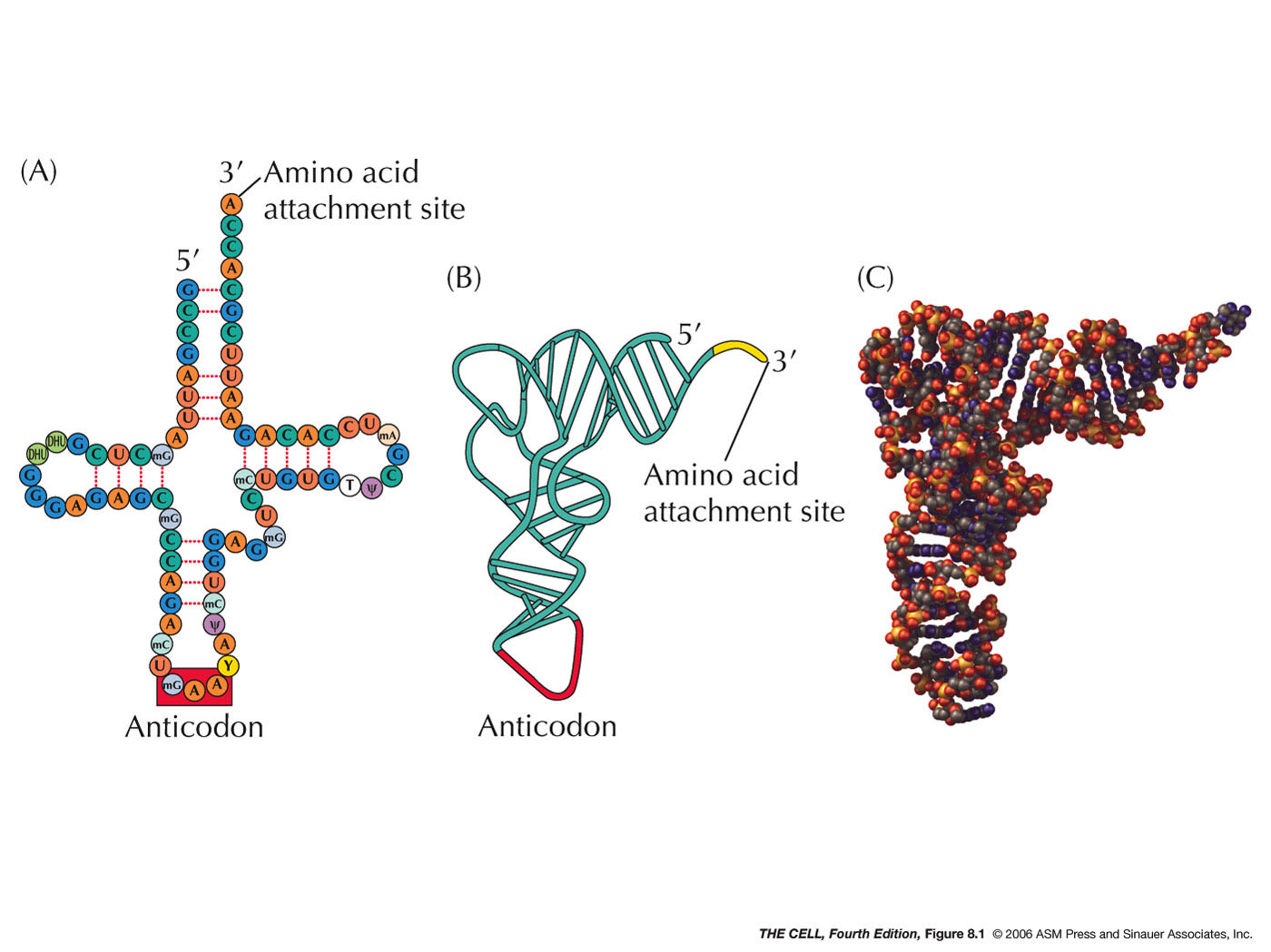 |
- tRNA's 3-D Structure: tRNAs all
have the same general shape, described as a
cloverleaf with internal base pairing holding the
cloverleaf in place. This cloverleaf actually
folds in on itself producing a more complex 3-D
structure.
|
- 3' CCA: All tRNAs have the
trinucleotide CCA at the 3' terminus (as we saw in
the last topic).
It is here that the amino acid will attach. The 3'
CCA is at one end of the folded tRNA and the
anticodon (see below) is at the other end.
|
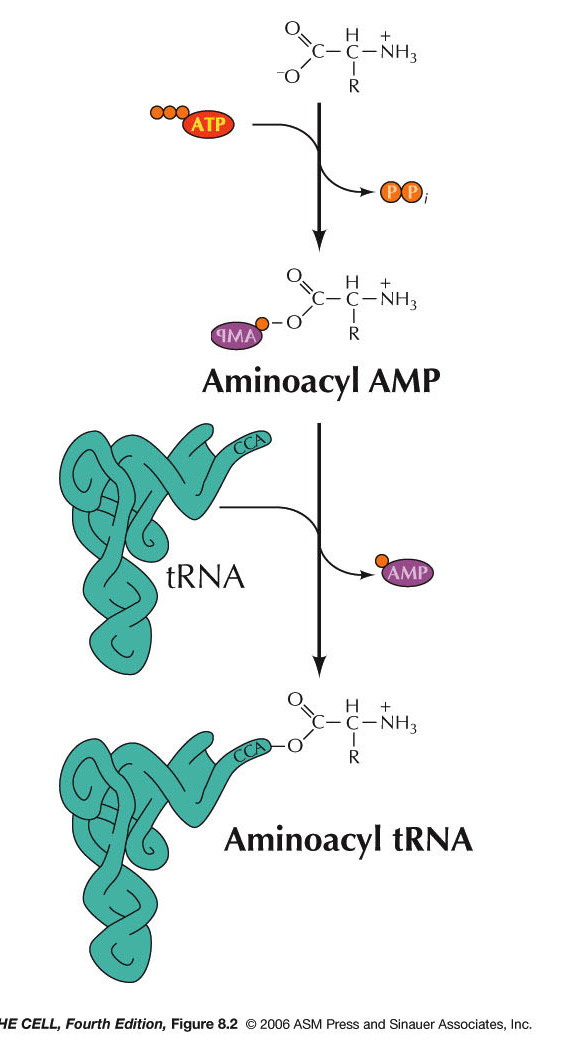
- Aminoacyl-tRNA Synthetases: The
attachment of an amino acid to the tRNA is
catalyzed by a group of enzymes called
aminoacyl-tRNA synthetases. There is a specific
enzyme for each amino acid/tRNA pair.
Aminoacyl-tRNA synthetase is the generic name,
with each amino acid having a specific enzyme. For
example, the enzyme that charges tRNAala
is alanyl-tRNA synthetase. A specific tRNA
synthetase is capable of binding specifically to
both the designated tRNA and to the designated
amino acid. This reaction requires ATP and occurs
in two steps. First,
tRNA synthetase activates the amino acid by adding
an AMP onto the carboxyl end of the amino acid
(using ATP and releasing pyrophosphate). The
carboxyl end is attached to the remaining
phosphate of the AMP. The amino acid is then
transferred to the oxygen at the 2' or 3' carbon
of the 3' end of the tRNA (the A nucleotide of the
CCA). (Type I and type II synthetases,
respectively.) Even if the amino acid is initially
attached to the 2' OH, it is the 3' OH form that
is used in protein synthesis.* This charged tRNA
is now ready to take part in translation. There is
one aminoacyl-tRNA synthetase for each amino acid
(20 of them) in most organisms. The one enzyme
recognizes more than one tRNA if there are cognate
tRNA for an amino acid. In bacteria, however,
there may be less than 20 aminoacyl-tRNA
synthetases**!
|
|
- Anticodon: Three bases in the
middle of the tRNA sequence make up the anticodon
(although they end up at one end of the tRNA after
it forms the cloverleaf and folds up). These three
bases are unique for each tRNA and (as we will
see) hydrogen bond to the codon of the mRNA during
translation. (See Degenerate below for the role of
modified bases in the anticodon.)
|
- The Ribosome: Ribosomes are the
sites where the actual protein synthesis occurs and
are similar in composition in prokaryotes and
eukaryotes. Like all large molecules and particles,
the size of ribosomes can be designated as a
sedimentation velocity. Prokaryotic ribosomes are
70S particles and eukaryotic ribosomes are 80S
particles. Ribosome are the smallest and the most
abundant of the organelles (20,000 in E. coli and as
many as 10 x 106 in some
eukaryotic cells). Ribosomes are composed of rRNA
and ribosomal proteins. The rRNAs have a 3-D
structure even more complex than that of the tRNAs
(rRNAs are much larger than tRNAs).
|
 |
- Prokaryotic Ribosomes: E. coli's
ribosomes are composed of a 50S and a 30S
subunit. The RNAs in the larger subunit are the
23S and 5S rRNAs (see last unit) and
it has 34 ribosomal proteins. The smaller
subunit has the 16S rRNA and 21 ribosomal
proteins.
|
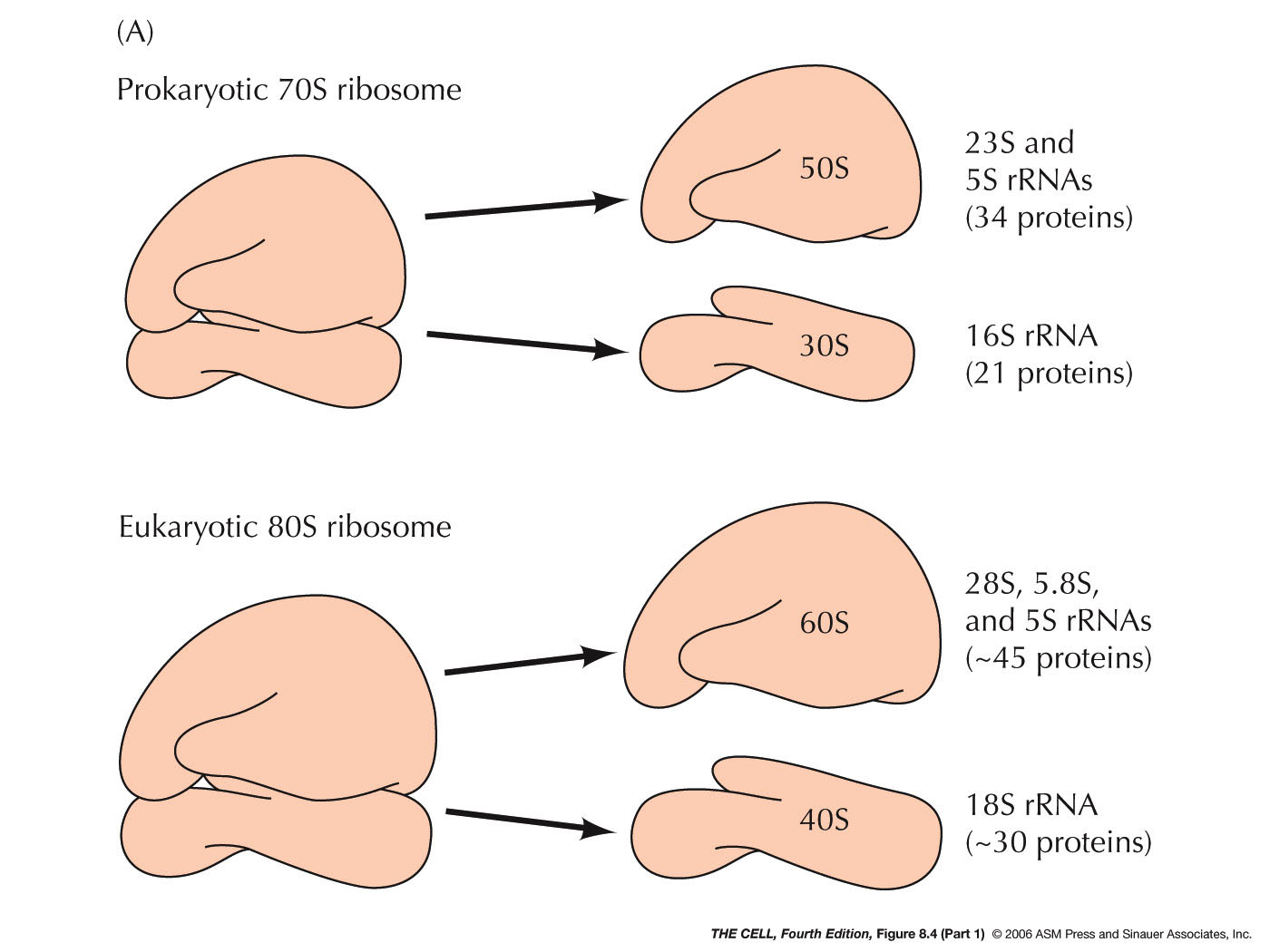 |
- Eukaryotic Ribosomes: The
subunits of a eukaryotic ribosome are slightly
larger than E.
coli's: the larger subunit is 60S and
the smaller one is 40S. The 60S subunit includes
the 28S, the 5.8S, and the 5S rRNA plus about 45
ribosomal proteins. The 40S subunit has the 18S
rRNA and about 30 ribosomal proteins.
- Ribosome Function: As we will see
below, the rRNAs and ribosomal proteins are
important in various binding and catalytic
activities of translation.
|
- mRNA: The mRNA molecule used in
translation has several features. (Remember, in
eukaryotes it has already undergone extensive processing, including
splicing.)
|
- Untranslated Regions: Even
after processing , mRNA molecules include a region
before the beginning of the coding sequence and
after the coding sequence ends. The "front" end of
the mRNA is referred to as the 5' untranslated
region (5' UTR) and the sequence at the "back" end
is called the 3' untranslated region (3' UTR).
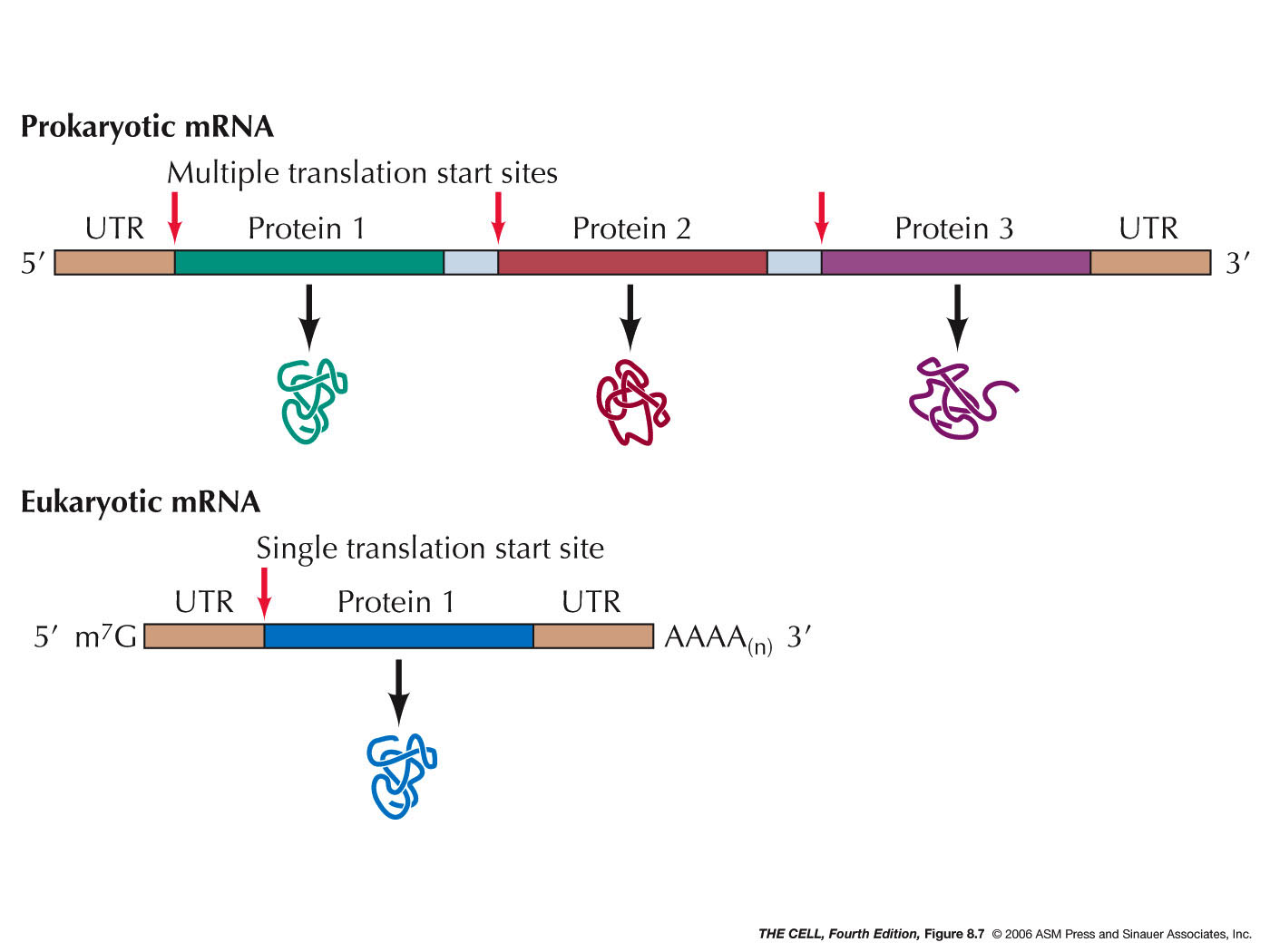
- Polycistronic
mRNAs: Prokaryotes often have long
mRNAs that code for more than one polypeptide.
These are referred to as polycistronic mRNAs.
Ribosome may dissociate from the mRNA at the end
of translation of a polypeptide, or stay on the
mRNA and continue translating the next CDS. (About
73% of E. coli's promoters control a
coding unit, meaning the rest control
polycistronic mRNA transcription.)(Some rare cases
of something similar occur in eukaryotes, but do
not use the same mechanism.)
|
 |
- Translation: The process of
translation is divided into these three stages:
|
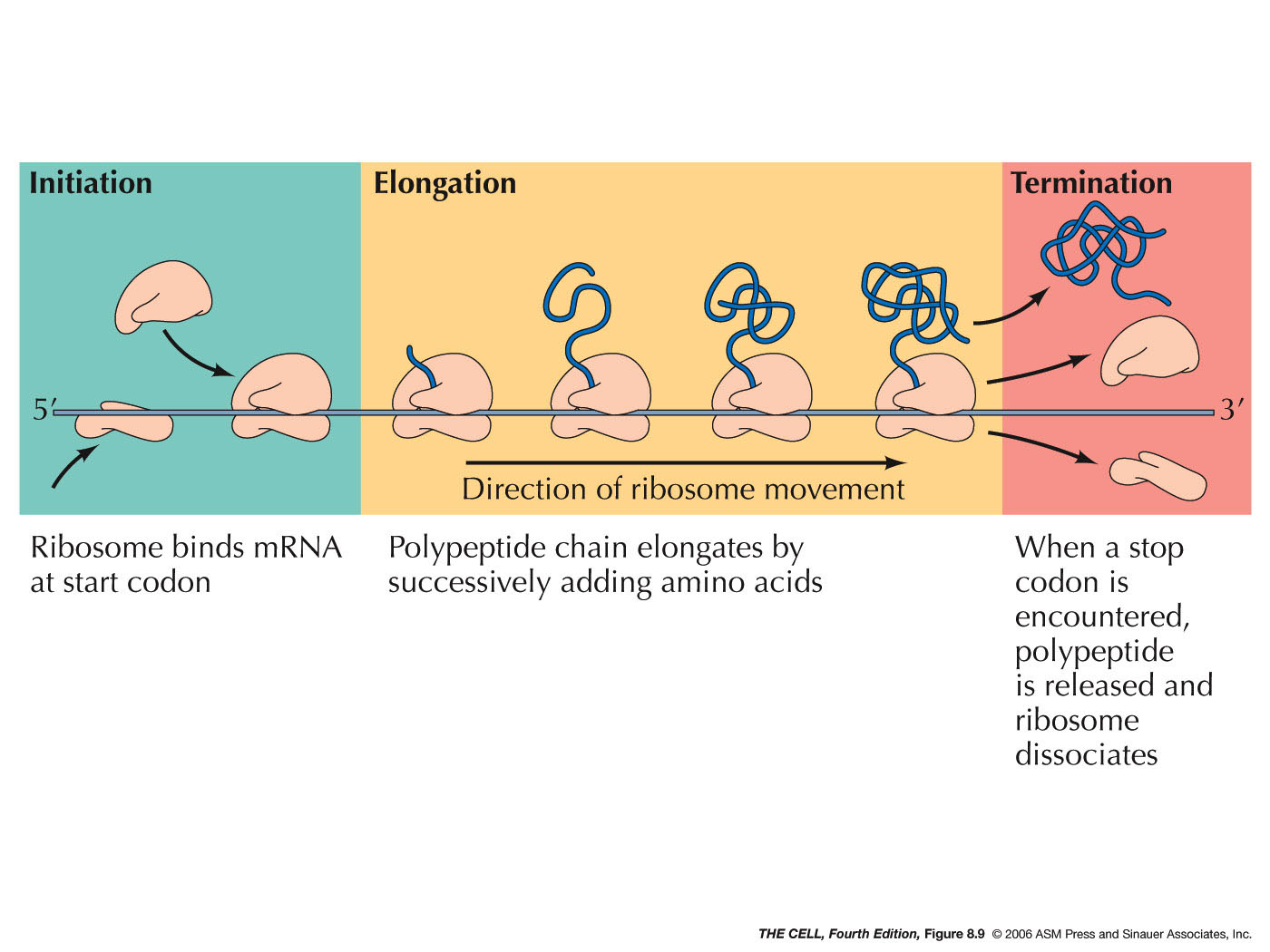 |
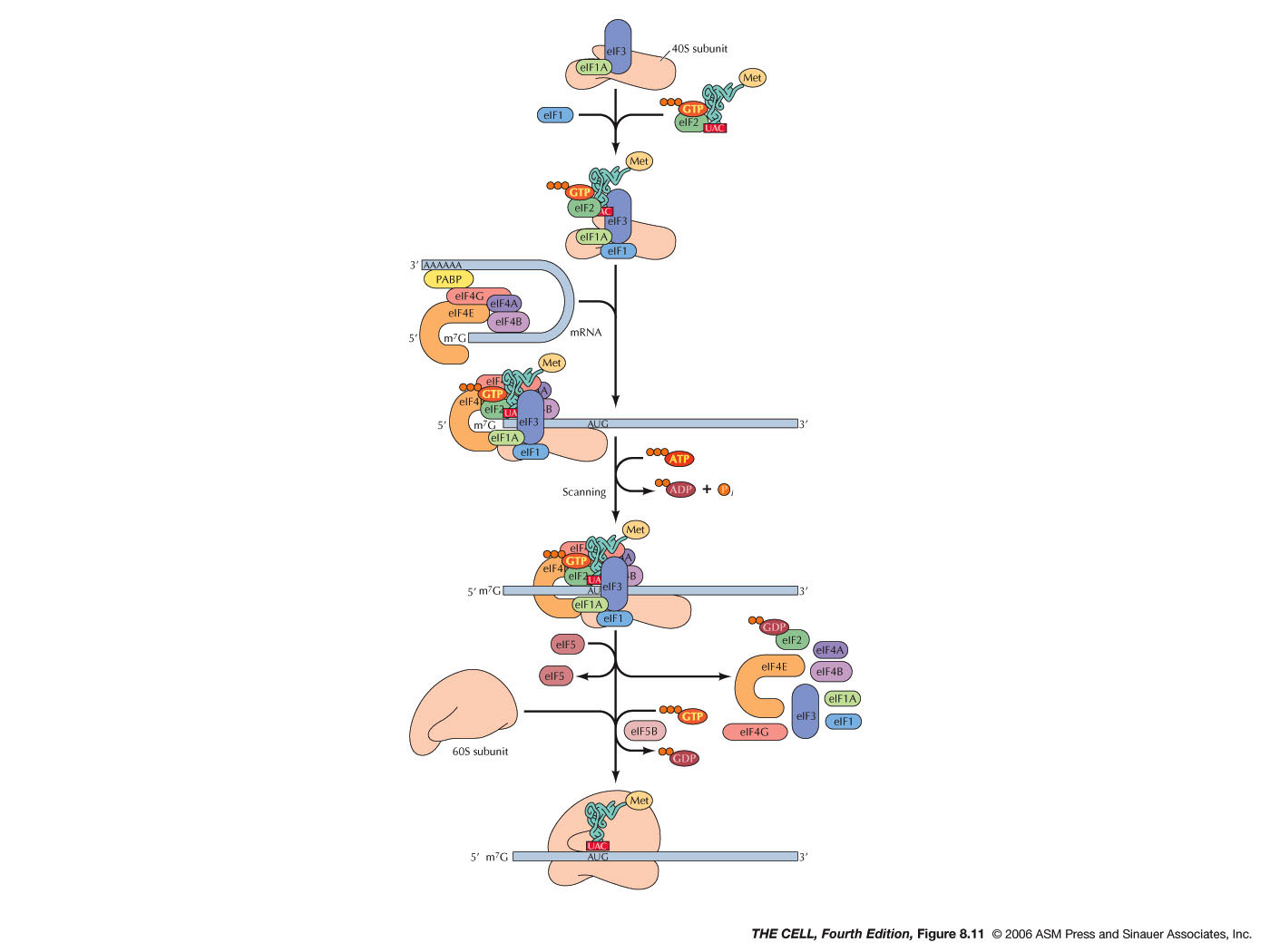
- Initiation:
During initiation in both prokaryotes and
eukaryotes, a special charged tRNA and the 5' end
of the mRNA bind to the small ribosomal
subunit. Initiation in both prokaryotes and
eukaryotes involves various protein factors.
|
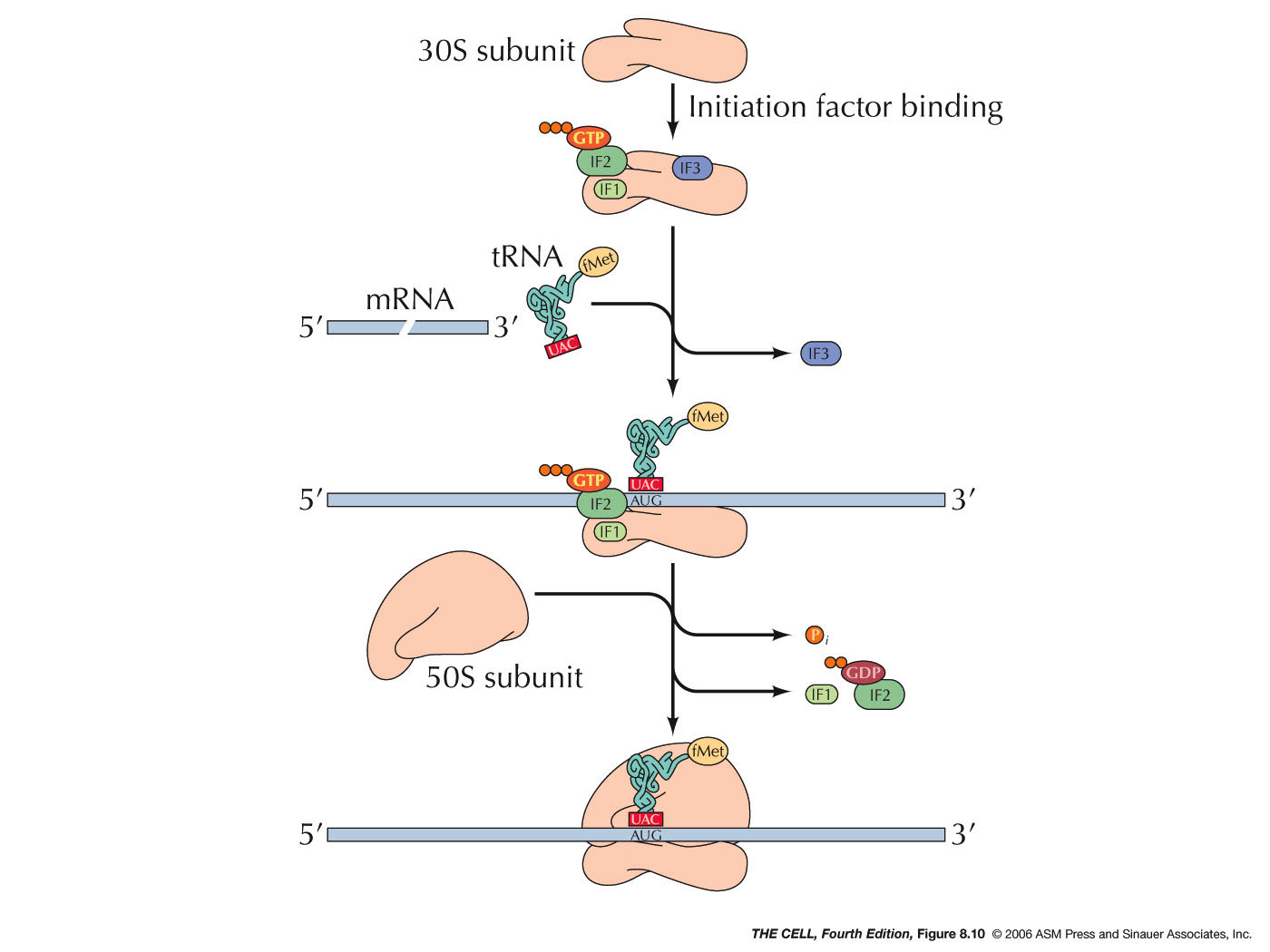
- Prokaryotes: Initiation in
prokaryotes begins with the binding of 3
initiation factors to the small subunit (IF2 has
GTP bound to it). Then, the mRNA and the special
charged tRNA bind to the complex. IF2 recognizes
this special tRNA. This tRNA is the initiator
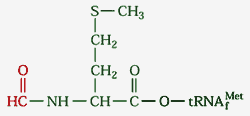
tRNA called tRNAfmet. It is charged
with a special amino acid: N-formyl
methionine (fmet-tRNAfmet).
This methionine has been modified by having a
formyl group (H-C=O) added to its N-terminus,
making it impossible to join with the C-terminus
of a previous polypeptide (only needed in
prokaryotes. Why?). A sequence (the Shine-Dalgarno
sequence: AGGAGG in most prokaryotes,
AGGAGGU in E. coli) on the mRNA is
recognized by a 16S rRNA sequence near its 3'
end. The ribosomal subunit then scans downstream
from this sequence on the mRNA until it finds
the AUG initiator codon (5-9 nucleotides
downstream), where translation will actually
begin. Since prokaryotes can have more than one
gene per mRNA, this means that the small
ribosomal subunit will also be able to recognize
an internal Shine-Dalgarno sequence and begin to
make a second (or third) polypeptide. The 50S
ribosomal subunit then joins and the IF2 is
released and its bound GTP is hydrolyzed to GDP
+ P and initiation is complete.
|
http://palaeos.com/bacteria/glossary/glossary.html#N-formyl-methionine
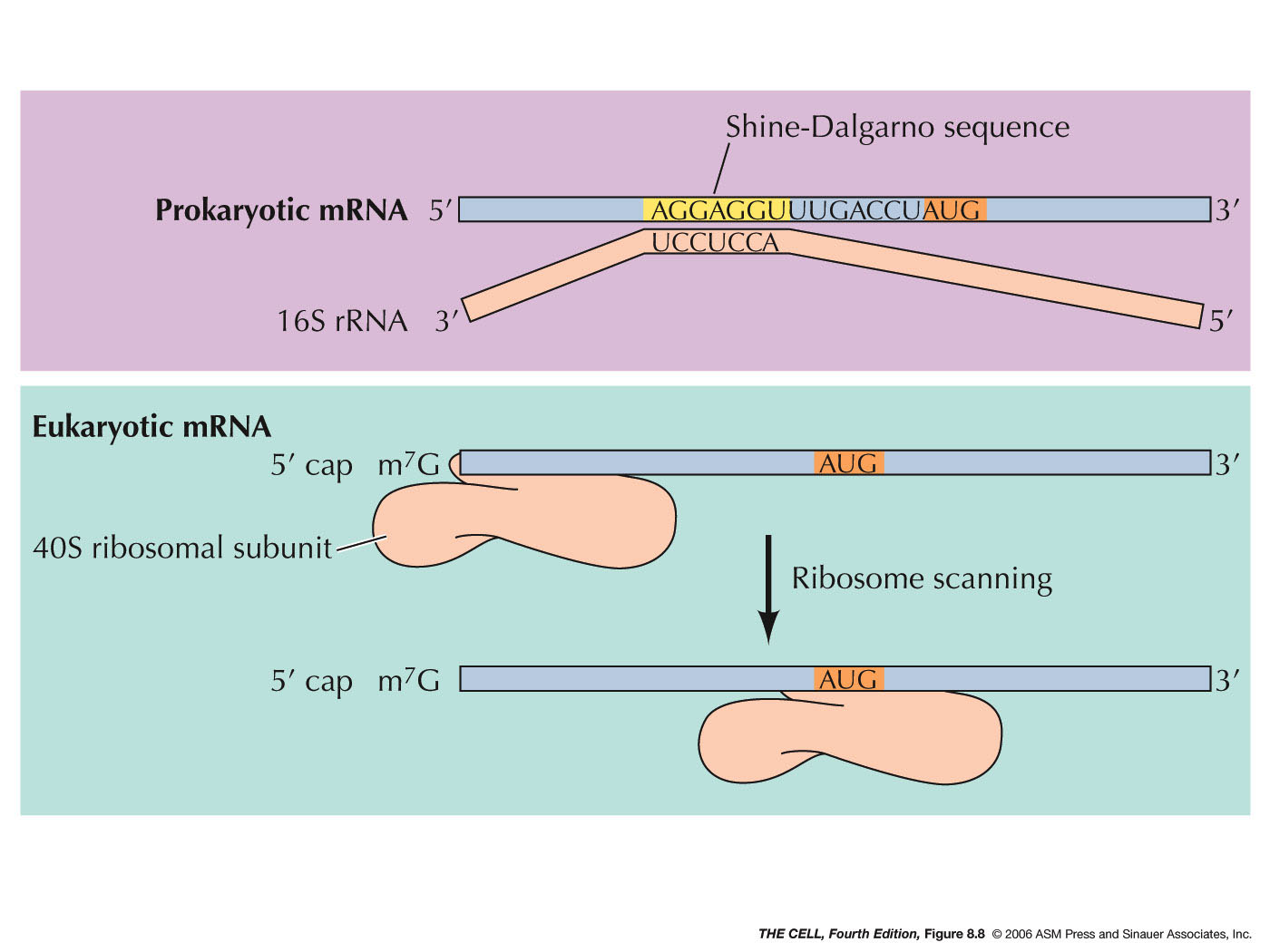 |
- Eukaryotes: During initiation in eukaryotes,
the tRNA and small ribosomal subunit binds to
the 5'
7-methylguanosine cap (instead of
the Shine-Dalgarno sequence) and scans from
there until it encounters the AUG initiation
codon. There are numerous protein factors
required for events like the binding to the
small ribosomal subunit, binding to the charged
initiator tRNA (in eukaryotes: met-tRNAmet), recognition
of the 5' cap, and recognition of the 3' poly-A
tail (eukaryotic initiation involves both the 5'
and 3' end of the mRNA and its folding). After
finding the AUG codon, the 60S subunits joins,
GTP is hydrolyzed and initiation is complete.
|
|
- Elongation:
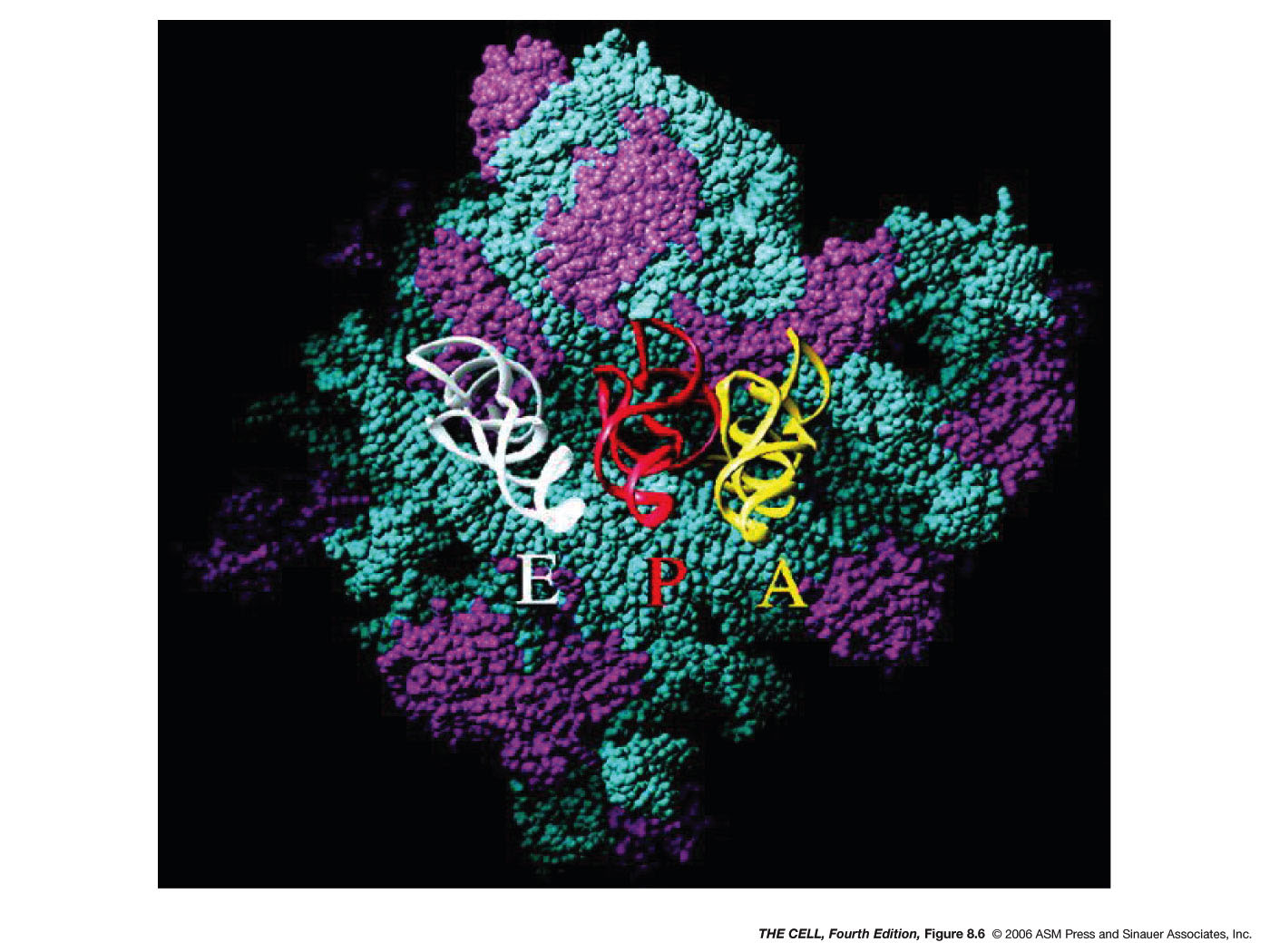
In both eukaryotes and prokaryotes, the assembled
ribosome has three potential tRNA binding sites:
the A site (aminoacyl
site), the P site (peptidyl site) and the E site
(exit site). The charged initiator tRNA
(fmet-tRNAfmet or met-tRNAmet)
is bound to the P site (which is between the other
two sites). Then, the next charged tRNA binds to
the A site. This binding is directed by the
codon-anticodon base pairing (antiparallelly
aligned). An RNA of the small ribosomal
subunit checks to make sure this pairing is
correct. Then, a GTP which was bound to an
elongation factor (there are many of them) is
hydrolyzed and that factor is released. Next, a
peptide bond is formed between the two adjacent
amino acids, with the amino acid in the P site
(its C-terminus is bound to the 3' end of its
tRNA) being transferred to the N-terminus of the
adjacent amino acid. The tRNA at the P site has
been released from its amino acid and the one at
the A site now has 2 amino acids attached to its
3' end. This catalytic activity is actually
carried out by RNA that is part of the 50S
subunit. Next, translocation occurs in which the
ribosome "moves" down the mRNA so that the tRNA
that was in the P site is now in the E site (it
exits) and the one that was in the A site is now
in the P site. This also requires GTP hydrolysis
and elongation factors. With the A site now open,
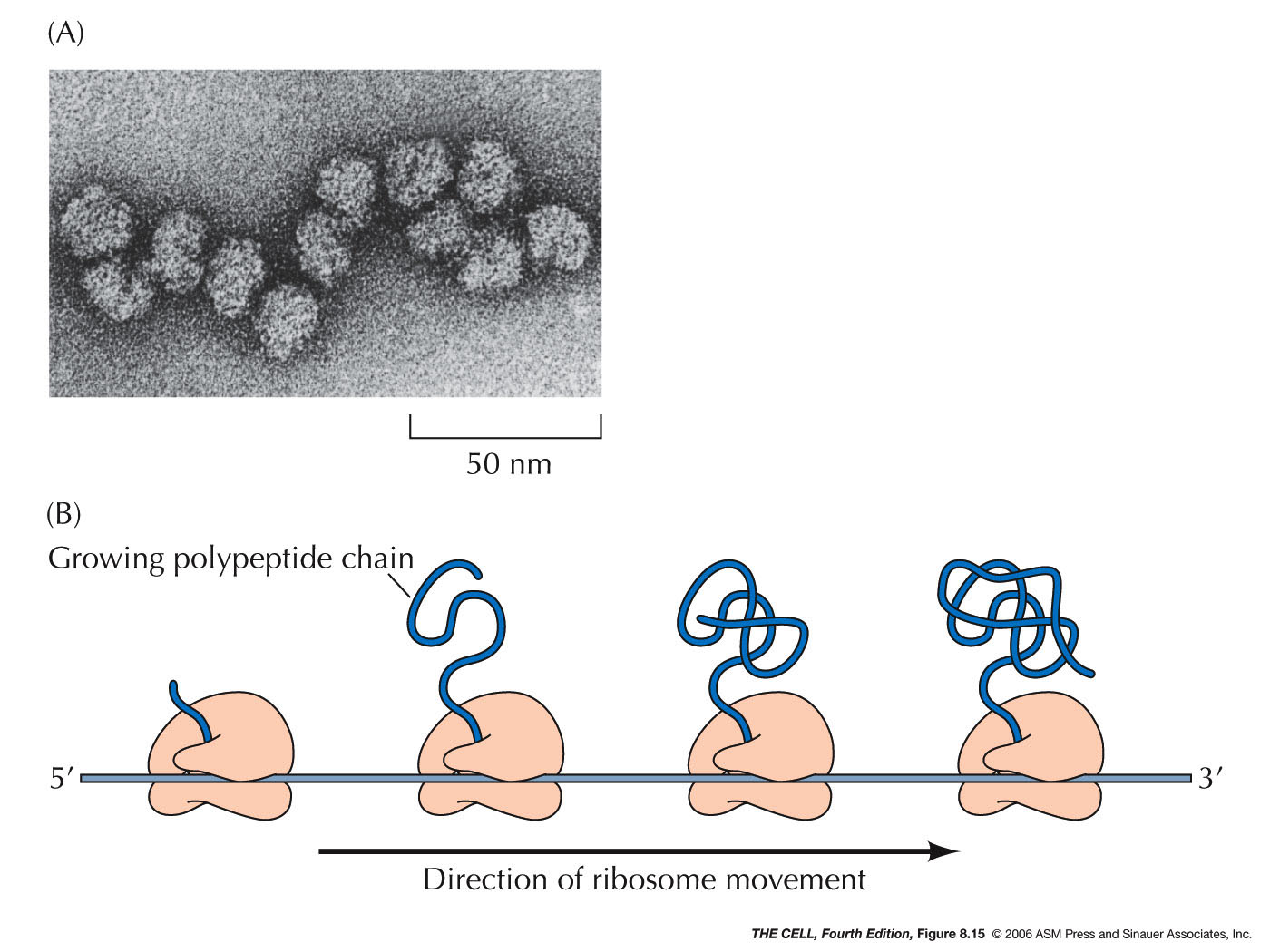
this process can be repeated and a polypeptide
made (15 amino acids/second). Several ribosomes (a
polysome or polyribosome)
may be translating a given mRNA at any time.
|
 |
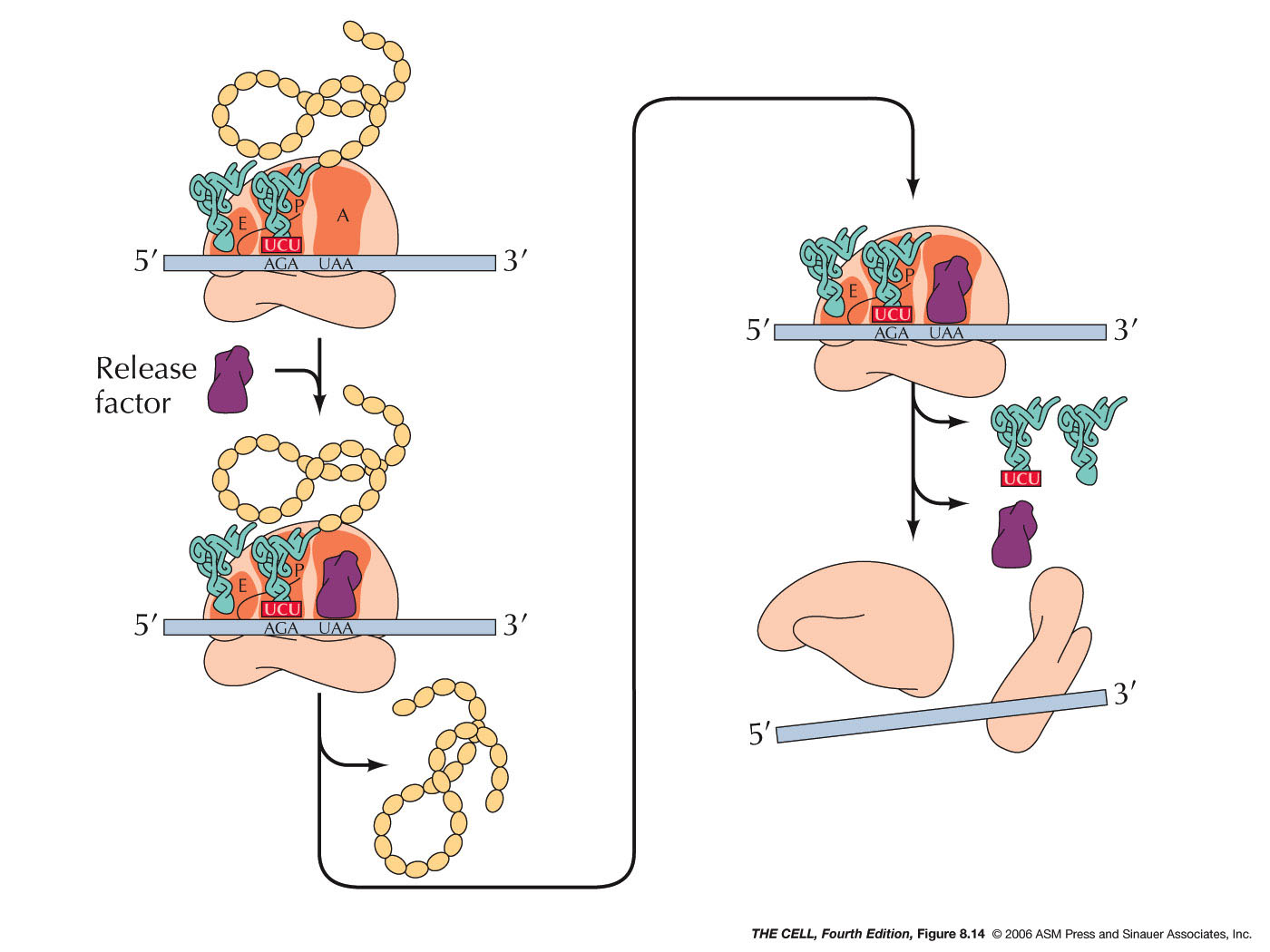
- Termination:
When a codon for which there is no tRNA with a
complementary anticodon comes into the A site,
translation terminates. These terminator codons
are UAA, UAG, and UGA (also called nonsense
codons). Instead of a tRNA binding to the open A
site, a release factor binds there and stops
protein synthesis. In prokaryotes there are two
release factors that do this work (one recognizes
UAA and UAG, the other recognizes UAA or UGA). In
eukaryotes, one factor recognizes all three
terminator codons. (A term you might run across:
ORF = open reading frame = a long DNA sequence
that could be transcribed into functional mRNA = a
potential "structural gene." That is, it has an
initiator codon and no terminator codon in the
same reading frame for some distance. Therefore
it could potentially be a polypeptide-coding
region.)
|
 |
|
|
 |
- Triplet: 3 bases per 1 amino acid
- Universal: The same code is used
in (almost)
all organisms.
- Non-ambiguous: A given codon
always encodes the same amino acid.
- Degenerate: More than one codon is
possible for most amino acids (64 codons, 20 amino
acids). This is accomplished primarily through "wobble" pairing at the
third codon position. (Wobble
pairing)(more)(more)
- Non-overlapping Well???
What about overlapping
genes.
- Commaless
(What about introns?)
|
*(From
"Aminoacylation of tRNA 2′- or 3′-hydroxyl by
phosphoseryl- and pyrrolysyl-tRNA synthetases,"
Published online 2013 Sep 8.
doi:10.1016/j.febslet.2013.08.037 by Englert et al.):
Aminoacyl-tRNA synthetases (AARSs) are
essential enzymes that catalyze the attachment of amino
acids to corresponding tRNAs [1]. The resulting
aminoacyl-tRNAs (AA-tRNAs) are transported by the
elongation factor (EF-Tu in bacteria and EF1A in archaea
and eukaryotes) to the ribosome as building blocks for
protein synthesis [2,3]. AARSs catalyze formation of
AA-tRNAs in two steps at the same active site:
activation of the amino acid with ATP to form an
aminoacyl-adenylate (AA-AMP), and transfer of the amino
acid moiety to the 2′- or 3′-hydroxyl group (OH) of the
tRNA terminal adenosine (A76) [1,4]. Based on the active
site structure, AARSs are grouped into two independently
evolved classes [5]. Class I AARSs all attach amino
acids to the 2′-OH of A76, whereas most Class II enzymes
aminoacylate the 3′-OH except asparaginyl- (AsnRS) and
phenylalanyl-tRNA (PheRS) synthetases that prefer the
2′-OH [4,6,7].
In solution, the 2′- or 3′-linked amino
acid spontaneously transacylates to the neighboring OH
at high rates [8], resulting in a mixture of 2′- and
3′-linked AA-tRNA isomers. EF-Tu stabilizes the
3′-isomer, which is preferred by the ribosome during
peptide bond formation [9]. The vicinal hydroxyl group
plays critical roles in catalyzing peptide bond
formation on the ribosome, and hydrolyzing (editing)
misacylated tRNAs by several AARSs and trans-editing
factors [10–13].
**(From Molecular Biology of the Cell, Alberts et
al., 5th ed., Garland Science): "Most cells have a
different synthetase enzyme for each amino acid (that
is, 20 synthetases in all); one attaches glycine to all
tRNAs that recognize codons for glycine, another
attaches alanine to all tRNAs that recognize codons for
alanine, and so on. Many bacteria, however, have fewer
than 20 synthetases, and the same synthetase enzyme is
responsible for coupling more than one amino acid to the
appropriate tRNAs. In these cases, a single synthetase
places the identical amino acid on two different types
of tRNAs, only one of which has an anticodon that
matches the amino acid. A second enzyme then chemically
modifies each 'incorrectly' attached amino acid so that
it now corresponds to the anticodon displayed by its
covalently linked tRNA."
|













 Home
Home Lectures
Lectures Videos
Videos Exams
Exams Extra
Extra